Harnessing AI to Accelerate Innovation in the Biopharmaceutical Industry
AI has the potential to transform drug development by enhancing productivity across the entire development pipeline, boosting biopharmaceutical innovation, accelerating the delivery of new therapies, and fostering competition to help improve public health outcomes.
KEY TAKEAWAYS
Key Takeaways
Contents
The Drug Development Process 3
The Role of AI in Drug Development 7
Challenges for AI Adoption. 19
Introduction
Drug development is a lengthy, complex, and costly process marked by extensive clinical trials, substantial risks, and stringent regulations. Artificial intelligence (AI) has the potential to transform the entire drug development process—from accelerating drug discovery and optimizing clinical trials to improving manufacturing and supply chain logistics.
An important challenge for innovation policy is to ensure that spending on medicines drives the greatest return to society. Policies that impose price controls or weaken intellectual property (IP) protections may aim to lower drug prices but are known to dampen incentives for future drug research and development (R&D) and fail to improve R&D productivity. A more sustainable approach is to pursue policies that maximize the value of R&D investment. Enhancing R&D productivity, especially in light of declining returns and rising risks that increase the cost of capital—a major factor in drug development costs—offers a more effective path to maximize public health returns, promote equitable access, and drive economic growth.
Technological advances hold promise for improving R&D productivity, accelerating access to novel therapies, and promoting health equity and competitiveness, ultimately delivering greater value for resources invested in medicines without stifling innovation. AI applications across various phases of drug development could enhance R&D productivity, increase drug output, and foster competition, driving innovation and expanding access to novel therapies to improve societal welfare. This increased drug supply can, in turn, drive consumer benefits through market competition. Furthermore, AI could accelerate the discovery of new therapeutic targets and drug candidates for the thousands of diseases that still lack treatment options.[1]
This report explores the potentially transformative role of AI across various phases of drug development, presenting early evidence highlighting how AI can enhance drug discovery, diversify clinical trials, and optimize drug manufacturing processes, ultimately leading to more efficient development with shorter timelines and improved outcomes. Additionally, the report identifies key challenges to broader AI adoption in drug development and offers several policy recommendations to support effective AI adoption, aimed at fostering innovation while ensuring patient safety.
The Drug Development Process
The drug development process is lengthy, risky, and costly.[2] While estimates vary widely across therapeutic areas, recent figures suggest that the R&D cost of bringing a new drug to market could be up to $2.83 billion (uncapitalized), factoring in pre- and post-approval R&D, such as new indications, patient populations, and dosage forms—or, capitalized at an annual rate of 10.5 percent, up to $4.04 billion.[3] According to a 2021 Biotechnology Innovation Organization (BIO) report, it takes more than a decade for a drug to reach the market, and only 7.9 percent of Phase I clinical trial candidates ultimately receive approval.[4]
Drug development is a complex, multiphase process that transforms scientific discoveries into safe, effective therapies. Key phases include drug discovery, preclinical testing, clinical trials, regulatory review, and manufacturing. (See figure 1.) In the drug discovery phase, scientists search for and identify therapeutic targets—such as proteins, including receptors and enzymes—involved in a disease. They then screen thousands of potential therapeutic compounds to find those capable of modulating the target’s activity. Promising compounds are refined and advanced to the preclinical testing phase, where lab and animal studies assess safety, efficacy, and pharmacokinetics/dynamics.[5]
The discovery and preclinical phases together typically take about five to six years.[6] After preclinical testing, these candidates move on to human clinical trials, which average 9.1 years.[7] Drugs that successfully complete clinical trials are submitted to regulatory authorities, such as the Food and Drug Administration (FDA), for approval based on evidence from both preclinical and clinical studies.[8] A recent report finds that, on average, it takes 10.5 years for a successful drug candidate to progress from Phase I clinical trials to regulatory approval, following years of early research to discover a therapeutic target and design a drug to effectively modulate it.[9]
Figure 1: The drug development process[10]
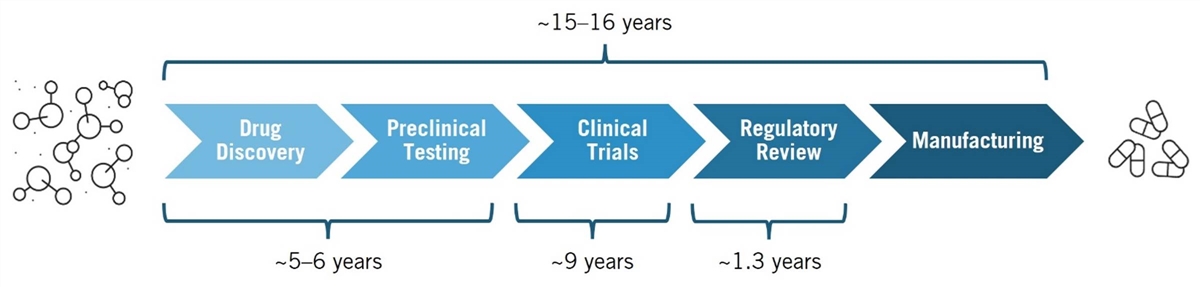 A prominent method in modern drug discovery is the target-based approach, which involves designing drugs that interact with specific therapeutic targets implicated in a disease. This method relies on a deep understanding of a target’s role in the disease to develop effective therapies. The process begins with identifying a target based on the hypothesis that modifying its activity will impact disease progression. Target identification involves analyzing extensive genomic, proteomic, and other biological data to pinpoint key molecules involved in disease pathways. Target identification is followed by validation through in vivo and ex vivo models. During this discovery phase, techniques such as high-throughput screening, bioinformatics, and experimental validation are employed to identify and confirm these targets. The process is laborious and complex, requiring significant time and effort. Once validated, drug candidates proceed to clinical trials to evaluate safety and efficacy in humans.[11]
A prominent method in modern drug discovery is the target-based approach, which involves designing drugs that interact with specific therapeutic targets implicated in a disease. This method relies on a deep understanding of a target’s role in the disease to develop effective therapies. The process begins with identifying a target based on the hypothesis that modifying its activity will impact disease progression. Target identification involves analyzing extensive genomic, proteomic, and other biological data to pinpoint key molecules involved in disease pathways. Target identification is followed by validation through in vivo and ex vivo models. During this discovery phase, techniques such as high-throughput screening, bioinformatics, and experimental validation are employed to identify and confirm these targets. The process is laborious and complex, requiring significant time and effort. Once validated, drug candidates proceed to clinical trials to evaluate safety and efficacy in humans.[11]
Drug development is a complex, multi-phase process that transforms scientific discoveries into safe, effective therapies. Key phases include drug discovery, preclinical testing, clinical trials, regulatory review, and manufacturing.
In practice, drug discovery often integrates target-based and phenotypic approaches. In phenotypic drug discovery, compounds are screened for their effects on disease-relevant traits (phenotypes) without prior knowledge of the exact molecular target or underlying mechanism. This method is particularly useful for complex conditions such as cancer and cardiovascular and immune diseases, which involve multiple genes and pathways. By combining these approaches, researchers can leverage their respective strengths to enhance the likelihood of developing effective therapies.[12]
In recent decades, advances in fields such as pharmacology, synthetic biology, and biotechnology—coupled with breakthroughs such as the decoding of the human genome in 2003 and the advent of next-generation sequencing techniques in the mid-2000s—have significantly expanded scientific understanding of health and disease. Over the past decades, biopharmaceutical R&D investments in the United States surged from $2 billion in 1980 to $96 billion in 2023.[13] However, despite the significant increase in resources, several indicators suggest that biopharmaceutical innovation is slowing (or plateauing).[14] Notably, there has as yet been no significant corresponding increase in the number of new drug launches, clinical trial success rates have decreased, and the overall length of drug development has increased. (See figure 2.)[15]
Moreover, over time, it has become more expensive to develop new drugs. A Deloitte report notes, “The average cost to develop an asset, including the cost of failure, has increased in six out of eight years.”[16] The 2019 version of the report concludes that the average cost of bringing a new biopharmaceutical drug to market has increased by 67 percent since 2010 alone.[17] At the same time, Deloitte found that forecast peak sales per asset have already more than halved since 2010. And significantly, the biopharma industry has experienced a downward trend in returns to pharmaceutical R&D: Deloitte found that the rate of return to R&D in 12 large-cap pharmaceutical companies declined from 10.1 percent in 2010 to 4.2 percent in 2015 and then to 1.8 percent in 2019.[18]
Figure 2: FDA new drug approvals, 1985–2023[19]
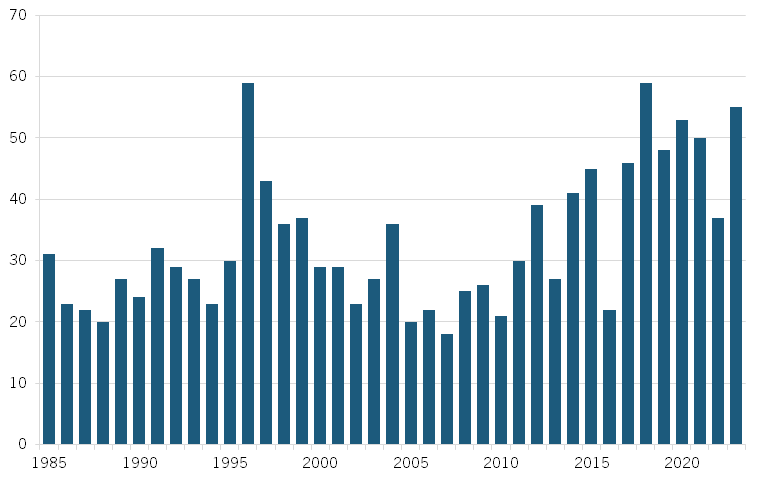
Several factors have been proposed to explain the decline in R&D productivity and the growing complexity of biopharmaceutical innovation.[20] These include shifts in the nature of science and technology, more stringent regulatory requirements, and a shift in R&D investments toward novel, high-risk, high-value targets that carry more uncertainty and difficulty.[21] Further, advancements in science, particularly in genetics, molecular biology, and systems biology, have revealed that many diseases are highly complex, resulting from a combination of genetic, environmental, and lifestyle factors. Such complexity often requires sophisticated treatments, such as targeted therapies or precision medicine, which require additional research and advanced technologies (e.g., gene editing, biomarker identification, etc.), adding time and cost to drug development.
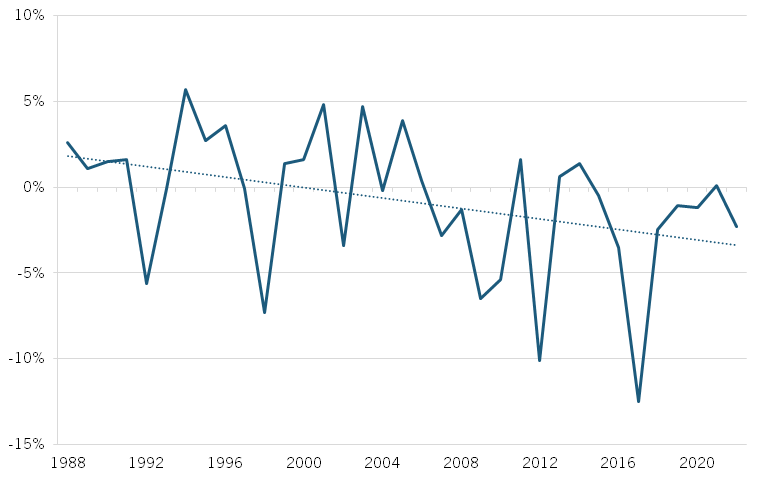
Beyond drug development—the process of discovering, designing, and testing new drugs— biopharmaceutical manufacturing, which involves producing drugs at scale, has also seen declining productivity. Total factor productivity (TFP), which measures the efficiency of all inputs (labor, capital, and materials) in production, decreased by nearly 2 percent annually from 2010 to 2018, indicating reduced output capacity. (See figure 3.) While TFP has since improved, growing at an average annual rate of 2 percent, labor productivity—measured by output per hour worked—has declined by more than 3 percent per year, pointing to an ongoing challenge. (See figure 4.) This decline could stem from factors such as stricter regulatory demands, a need for more specialized handling and monitoring, or greater manufacturing complexity, which may be reducing labor input efficiency, even if overall TFP has improved. These factors are likely linked to the industry’s growing pursuit of more advanced treatments, such as cell and gene therapies, which require more intricate handling and resource-intensive manufacturing processes.
Figure 3: Annual change in biopharmaceutical manufacturing TFP, 1988–2021[22]
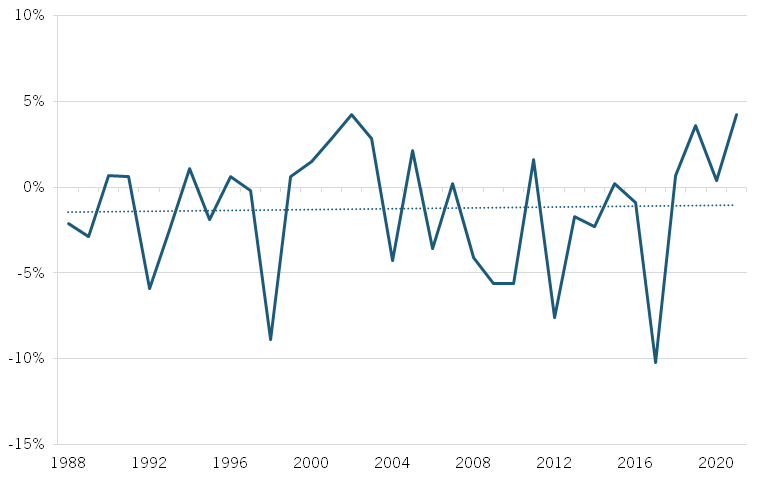
Figure 4: Annual change in biopharmaceutical manufacturing labor productivity, 1988–2021[23]
The Role of AI in Drug Development
Amid declining productivity in drug development, emerging technologies offer the potential to boost biopharmaceutical productivity. Advancements such as AI, quantum computing, CRISPR (clustered regularly interspaced short palindromic repeats), 3D bioprinting, organ-on-a-chip, and nanotechnology could significantly reduce the time and cost of bringing new therapies to market, making life-saving therapies more widely accessible to patients. Next-generation sequencing has enabled the production of large biomedical datasets, particularly genomics (the study of an organism’s genes) and transcriptomics (the study of RNA transcripts), among others.[24] Along with data from experimental research and pharmaceutical studies, these vast datasets provide fertile ground for AI, which can analyze these datasets to uncover patterns and make predictions that serve as valuable inputs across different phases of drug development—from target identification and clinical trials to regulatory processes, manufacturing, and supply chain optimization.
Quantum computing, while still in the early stages, can complement AI by solving problems beyond the capabilities of classical computers—such as molecular simulations and protein folding—both critical for modeling biological systems and identifying new drug candidates. Together, these technologies can accelerate biopharmaceutical innovation, offering more efficient pathways to novel therapies.[25]
In Prediction Machines: The Simple Economics of Artificial Intelligence, Ajay Agrawal, Joshua Gans, and Avi Goldfarb described AI as a tool that transforms data into predictions, offering valuable insights to inform decision-making. In the biopharmaceutical industry, AI can analyze vast biological datasets to predict which therapeutic targets are linked to diseases, identify promising drug candidates, and forecast drug responses.[26] Using advanced machine learning techniques, particularly deep learning based on neural networks trained on large datasets, AI excels at recognizing patterns and generating predictions. Importantly, AI complements rather than replaces human scientists. A key AI limitation lies in performing advanced causal inference —an essential task that requires human judgment. While AI is adept at identifying correlations, such as linking a target to a disease, it struggles to grasp the complex underlying biological mechanisms that explain why these links exist—a crucial aspect of effective drug development.
The vast amount of data provides fertile ground for AI, which can analyze datasets to uncover patterns that support each phase of drug development, from target identification and clinical trials to regulatory processes, manufacturing, and supply chain optimization.
Still, AI provides a powerful tool that can significantly advance different phases of drug development, from discovery and preclinical testing to clinical trials, regulatory review, and manufacturing. In drug discovery, AI accelerates the analysis of large datasets to identify promising compounds, reducing the time and cost of finding new drug candidates. During preclinical testing, AI models simulate biological processes to predict how a drug will behave in humans, reducing reliance on animal testing. In clinical trials, AI can optimize patient selection, improve trial design, and speed up data analysis, leading to faster, more accurate outcomes. It can also streamline the regulatory process by assisting in the preparation of complex documentation required by agencies such as America’s FDA and Europe’s European Medicines Agency (EMA). Finally, in manufacturing, AI can enhance production processes, improving efficiency and ensuring consistent quality. By integrating AI throughout these stages, the process of bringing a new drug to market can become more efficient, possibly bringing much-needed therapies to market quicker.
Facilitating Drug Discovery
AI is often hailed as the future of drug discovery, with the ability to reduce the time and cost of identifying therapeutic targets and promising drug compounds, optimize drug chemical structures for greater efficacy, and enhance the molecular diversity of potential targets.[27] Traditional experimental methods for target discovery can be slow and limited in scope, but AI offers a more efficient approach by predicting a set of targets from a large dataset based on properties that suggest their involvement in disease.[28] Scientists can then validate these predictions and develop therapeutic hypotheses about why those targets are linked to disease in order to develop safe and effective therapies.[29]
Illustrative Example: Identifying Therapeutic Targets in Lung Cancer
AI is transforming drug discovery by helping scientists identify potential therapeutic targets in cancer by analyzing vast genomic and clinical data.
Goal: Suppose a lung cancer research team aims to identify new therapeutic targets to guide drug development. The team plans to use AI to predict genes associated with lung cancer that could serve as therapeutic targets. If a gene consistently shows higher expression levels in cancer patients, it could represent a promising target for developing a novel drug.
Data: A dataset of 10,000 individuals, including 5,000 with lung cancer (labeled as 1) and 5,000 without (labeled as 0). Contains data on each individual’s gene expression levels for 20,000 genes, obtained through whole-genome sequencing.
The Role of Human Scientists: Before training the AI model, researchers apply their deep knowledge of biological processes and causal inference to narrow the list of genes from 20,000 to a more relevant subset, say 500 genes. This selection process often involves formulating hypotheses regarding the genes’ causal role in cancer development, employing causal inference methods and reviewing existing scientific literature to identify genes linked to lung cancer progression, drug resistance, and treatment outcomes. By focusing on biologically relevant genes, scientists enhance the likelihood of identifying meaningful drug targets while managing the complexities of high-dimensional genomic data. This approach improves the predictive power of AI models and ensures that the selected genes have a solid biological foundation.
The Role of AI: The AI model employs predictive modeling through statistical models—such as logistic regression, probabilistic regression, bagging (e.g., random forest), boosting, and others— which predict whether a patient has lung cancer based on gene expression levels. The models can identify patterns in the data, assign weights to each gene, and test the accuracy of its predictions on partitions of the dataset.[30]
How It Works: Training the AI model involves analyzing gene expression levels for the subset of 500 genes across the 10,000 individuals and looking for correlations with lung cancer status. AI models assign a weight to each gene, indicating the extent to which each gene contributes to predicting the likelihood of lung cancer. Genes with large positive weights—or higher importance—are flagged as potential drug development targets.
In July 2021, Google DeepMind’s AlphaFold2 system solved a pivotal aspect of the long-standing “protein-folding problem,” a 50-year-old biology challenge. By predicting the 3D structures of nearly all known proteins from their amino acid sequences, AlphaFold2 has transformed a critical field of biological research. The system, using a vast database of known protein structures, reduced the time needed to predict protein structure from months to minutes. In collaboration with the European Molecular Biology Laboratory (EMBL), Google DeepMind made this data freely available to the scientific community.[31] This breakthrough has profound implications for understanding protein function, which is key for designing more effective drugs.[32] This achievement earned Demis Hassabis and John M. Jumper of Google DeepMind the 2024 Nobel Prize in Chemistry “for protein structure prediction.” The prize was also shared with David Baker “for computational protein design.”[33]
AI is often hailed as the future of drug discovery, with the ability to reduce the time and cost of identifying therapeutic targets and promising drug compounds, optimize drug chemical structures for greater efficacy, and enhance the molecular diversity of potential targets.
Additionally, AI can assist with bioactivity prediction—evaluating how effectively a drug interacts with its intended target—by identifying promising compounds from large candidate pools, thereby minimizing the need for costly, time-consuming experiments. In 2024, a research team developed ActFound, an AI model that employs pairwise and meta-learning techniques, trained on millions of data points from ChEMBL, a public bioactivity database maintained by EMBL’s European Bioinformatics Institute (EMBL-EBI). ActFound has been shown to be more accurate and less costly than traditional computational methods.[34]
Genentech’s Lab-in-the-Loop
Genentech, a biotechnology pioneer founded in 1976 and now part of the Roche Group, is dedicated to combating serious, life-threatening diseases. Its notable achievements include the development of the first targeted antibody for cancer and the first drug for primary progressive multiple sclerosis.
In 2023, Genentech partnered with NVIDIA to launch an AI platform named “lab-in-the-loop,” designed to accelerate the use of generative AI in drug discovery and development. Created by Genentech’s Prescient Design accelerator unit, the platform utilizes Genentech’s granular data to train algorithms for designing new drug compounds. These compounds are tested in the lab, and the resulting data is fed back to refine the AI algorithms. This interdisciplinary approach creates a continuous loop between wet lab work and computational methods, evolving experimental and clinical data into predictive models for potential drug candidates, thus accelerating the development of life-saving therapies.[35]
Dr. Aviv Regev, executive vice president and head of Genentech Research and Early Development, described the lab-in-the-loop approach as:
the mechanism by which you bring generative AI to drug discovery and development. When we try to discover drugs, we’re only as good as our data. We take the data and we use it to train algorithms; use these algorithms that we’ve trained to generate new kinds of molecules that we haven’t tested before, which we will take back to the lab, and generate experimental data for them again. And those test results will be sent to the AI, to improve itself, to get a better algorithm, and we repeat this process again and again and again until we reach a molecule that has all the right properties that we need for it to be a real medicine for patients.[36]
In January 2024, Genentech began recruiting patients for a clinical trial to test the effectiveness of its experimental drug vixarelimab in treating ulcerative colitis, an inflammatory bowel disease. The drug had previously been tested only in lung and skin disorders. Traditionally, determining a drug’s potential for different indications can take years of laboratory work, but Genentech’s AI platform expedited this process, helping scientists determine in just nine months that vixarelimab could be a promising drug candidate for treating diseases affecting colon cells.
Genentech leverages AI in drug discovery to deepen scientists’ understanding of health and disease, using these insights to develop more effective therapies. By integrating knowledge from fields such as biology, computation, genomics, and machine learning, Genentech aims to drive biopharmaceutical innovation.
While AI is still in its early stages, several indicators suggest that it is already streamlining drug discovery. A study by the Boston Consulting Group (BCG) examines the research pipelines of 20 AI-focused pharmaceutical companies and finds that 5 out of 15 AI-assisted drug candidates that advanced to clinical trials did so in under four years, compared with the historical average of five to six years.[37] A 2023 report by BCG and the Wellcome Trust projects that AI-enabled efforts could reduce the time and cost of the drug discovery and preclinical stages by 25 to 50 percent.[38] And a 2019 report by the U.S. Government Accountability Office and the National Academy of Medicine notes that one company estimated AI-accelerated drug discovery could save between $300 million and $400 million per drug—stemming from improved R&D productivity, as AI increases the efficiency of capital investment, enabling better drugs to be identified earlier and more quickly.[39]
AI can significantly advance the main phases of drug development—from discovery and preclinical testing to clinical trials, regulatory review, and manufacturing.

Several AI-enabled drug discovery companies have begun reporting significantly accelerated drug discovery timelines. In September 2019, Canadian biotechnology firm Deep Genomics announced its first AI-discovered therapeutic drug candidate aimed at treating Wilson’s disease, a genetic disorder that leads to excess copper in the blood, causing liver and neurological issues.[40] This drug candidate was proposed just 18 months after target discovery efforts began, with the company’s AI platform analyzing over 2,400 diseases and 100,000 pathogenic mutations.[41] In January 2020, British pharmaceutical company Exscientia, in collaboration with Japan’s Sumitomo Dainippon Pharma, reported that its AI-developed compound for obsessive-compulsive disorder (OCD) had reached clinical trials in just 12 months, compared with the typical five- to six-year timeline. This compound was also reportedly the first AI-designed drug to enter clinical trials.[42] In June 2023, Hong Kong- and New York-based Insilico Medicine announced that it had advanced its AI-developed drug for idiopathic pulmonary fibrosis, a chronic lung disease, to clinical trials in under 30 months.[43] While this early evidence suggests that AI has the potential to significantly reduce the speed of drug discovery, it will take longer to determine how AI-developed drugs perform in clinical trials compared with their non-AI counterparts.[44]
The BCG study reveals that AI-enabled drug discovery companies tend to concentrate their pipelines on well-established therapeutic target classes.[45] Over 60 percent of their disclosed targets come from familiar classes, such as enzymes (e.g., kinases) and G-protein-coupled receptors. This focus on well-known targets likely reflects a strategy to mitigate the risks associated with drug development and to demonstrate the viability of their AI platforms. In contrast, large pharmaceutical companies typically maintain more diverse pipelines, balancing both new and established targets.[46]
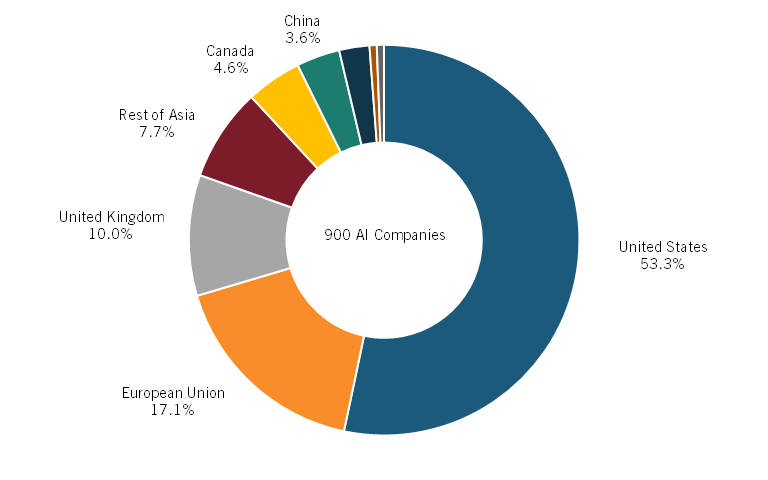
Moreover, a recent report from Deep Pharma Intelligence shows that in the geographical landscape of AI adoption in drug discovery, the United States is a global leader. As of 2023, more than 50 percent of all AI-enabled drug discovery biotechnology companies were based in the United States, followed by 17 percent in Europe and close to 4 percent in China. (See figure 5.)[47]
Figure 5: Share of companies using AI for drug discovery, 2023[48]
Moreover, the number of partnerships between large pharmaceutical companies and AI companies has surged, from 21 such new partnerships in 2017 to 66 in 2022, a more than threefold increase. (See figure 6, reproduced from the report.)[49] According to a recent study by S&P Global Ratings, examples of such partnerships in drug discovery include AstraZeneca and BenevolentAI, which have partnered for target identification optimization; GSK and Insilico Medicine, which are working together on the identification of novel biological targets and pathways; and Pfizer and Genetic Leap, which are pursuing the development of RNA genetic drug candidates.[50]
Figure 6: Number of AI-focused partnerships for large pharma companies, 2017–2022[51]
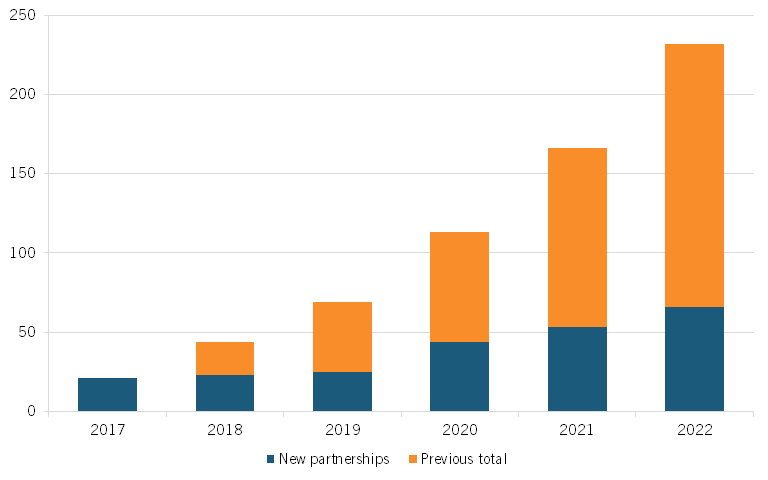
Streamlining Clinical Trials
Clinical trials, used to assess the safety and efficacy of proposed new therapies, represent a crucial step in the drug development process. However, conducting them is expensive, time consuming, and risky.[52] As of 2020, the global clinical trial market was valued at $44.3 billion.[53] One study finds that the average clinical development time for a typical drug—from first-in-human trials to regulatory approval—is 9.1 years, though this can vary by therapeutic area, indication, trial design, and patient availability.[54] A recent BIO report details that, on average, Phase I trials take 2.3 years, Phase II 3.6 years, Phase III 3.3 years, and regulatory review 1.3 years.[55] Furthermore, fewer than 8 percent of drug candidates that enter Phase I trials succeed.[56] Given these challenges, AI has the potential to streamline and improve clinical trials in several ways, making them faster and more effective and cost-efficient.[57]
Enhancing Patient Eligibility and Recruitment
One of the most time-consuming aspects of clinical trials is identifying and recruiting eligible patients, a process that can take up to one-third of the total trial duration. Moreover, 20 percent of trials fail to recruit the required number of participants. To address this challenge, researchers have explored relaxing overly strict eligibility criteria while maintaining patient safety.[58] Trial Pathfinder, an AI system developed at Stanford University, analyzes completed clinical trials to assess how modifying criteria, such as blood pressure thresholds, impacts adverse events such as serious illness or death among participants.[59] In a study of non-small cell lung cancer trials, Trial Pathfinder showed that loosening certain criteria—some of which, like lab test results, had little impact on the trial’s outcome—could double the number of eligible patients without increasing the risk of adverse events. This approach proved effective for other cancers, including melanoma and follicular lymphoma, and in some cases, even reduced negative outcomes by including sicker patients who stood to benefit more from the treatment in the trial.[60] These findings suggest that relaxing restrictive eligibility criteria can expand patient access to promising new therapies.
After determining eligibility criteria, the next major challenge is recruiting patients, as failure to do so can produce delays and trial terminations.[61] Typically, patient recruitment involves manual prescreening due to the complexity of clinical criteria text, which often includes confusing abbreviations and terminology.[62] Criteria2Query is an AI system that can streamline this process by parsing eligibility criteria from natural language and converting it into structured, searchable data. Researchers can input inclusion and exclusion criteria in plain language that Criteria2Query translates into formal database queries that sift through electronic health records (EHRs) to find matching participants.[63] This system aims to enhance human–AI collaboration, optimizing cohort generation by combining machine efficiency with human expertise to simplify complex concepts and train algorithms. Criteria2Query can accelerate recruitment and help include populations such as children and the elderly, who are often unnecessarily excluded from trials, thereby speeding up and diversifying clinical trials.
Several AI-enabled drug discovery companies have begun reporting significantly accelerated drug discovery timelines.
Beyond systems such as Criteria2Query, which match trials to patients, AI also improves patient-to-trial matching. TrialGPT, for example, helps patients find suitable trials by predicting their eligibility for specific studies. Patients provide a description of their condition in plain language, and TrialGPT generates a score reflecting their fit for a given trial, along with explanations for each eligibility criterion. In testing, has TrialGPT achieved nearly human-level accuracy and reduced screening time for trial matching by over 40 percent, highlighting the potential of AI to enhance clinical trial efficiency and accelerate patient recruitment.[64] In October 2023, the Dana-Farber Cancer Institute, supported by a Meta grant, began developing a novel open source AI platform to “computationally match patients with cancer to clinical trials.”[65] This initiative is leveraging Meta’s large language model, Llama 3, to analyze unstructured clinical notes and trial eligibility criteria, enabling quicker and more accurate matching of patients to suitable clinical trials.[66]
AI can also play a critical role in identifying biomarkers that predict therapy outcomes and disease progression, which is crucial for matching patients to the most effective clinical trials and therapies. A research team led by Dan Theodorescu, director of Cedars-Sinai Cancer in Los Angeles, developed the Molecular Twin Precision Oncology Platform (MT-POP) to discover biomarkers that predict disease progression in pancreatic cancer, one of the most lethal cancers. Their findings revealed that relying solely on CA 19-9—the only FDA-approved biomarker for pancreatic cancer—was suboptimal for predicting therapy outcomes.[67] Instead, a set of multiple biomarkers provided far more accurate predictions. This multi-omics approach demonstrates the potential to optimize patient selection for clinical trials—as well as approved therapies—by identifying those most likely to benefit from specific treatments.[68] Such advancements could help refine trial designs and improve the likelihood of success in developing new therapies for pancreatic ductal adenocarcinoma and other challenging diseases.
Optimizing Clinical Trial Design
AI can also enhance clinical trial design by optimizing aspects such as drug dosages, patient enrollment numbers, and data collection strategies. At the University of Illinois, researchers developed the HINT (Hierarchical Interaction Network) algorithm to predict trial success based on factors such as the drug molecule, target disease, and patient criteria. Trained on pharmacokinetic and historical trial data, HINT has demonstrated high accuracy across different clinical trial phases and diseases.[69] Building on this foundation, the team created SPOT (Sequential Predictive Modeling of Clinical Trial Outcome), which tracks trial progress in real time and refines predictions as more data becomes available, allowing for timely adjustments during the trial to improve outcomes.[70] Moreover, the Illinois-based company Intelligent Medical Objects has developed SEETrials, which utilizes OpenAI’s GPT-4 to extract safety and efficacy data from clinical trial abstracts, helping clinical researchers evaluate different trial designs.[71]
Further, AI-powered clinical trial design start-up QuantHealth has developed Katina, an AI platform that simulates hundreds of thousands of potential trial protocol combinations—such as various patient groups, treatment parameters (e.g., treatment dose, administration route, duration), and trial endpoints (e.g., tumor shrinkage)—to enhance the likelihood of trial success. Trained on extensive biomedical, clinical, and pharmacological data, the AI-guided workflow aims to enhance trial design and execution.[72] In August 2024, QuantHealth reported that Katina had simulated over 100 trials with 85 percent accuracy. It could predict Phase II trial outcomes with 88 percent accuracy, significantly higher than current success rates of 28.9 percent, and Phase III trial outcomes with 83.2 percent accuracy—compared with 57.8 percent. The platform was also able to reduce trial costs and durations. QuantHealth reported that its collaboration with a pharmaceutical company’s respiratory disease team led to a significant $215 million reduction in clinical trial costs, achieved by shortening trial duration by an average of 11 months, requiring 251 fewer trial participants, and using 1.5 fewer full-time employees to conduct the trial.[73]
Streamlining Clinical Trial Protocols
AI can also streamline the analysis of clinical trial protocols (CTPs), which are typically lengthy, complex documents exceeding 200 pages. CTPs cover everything from trial objectives and design to participant eligibility criteria and statistical methods, serving as a blueprint to conduct trials in a structured, compliant way. By using AI to extract valuable insights from unstructured documents, researchers can enhance participant diversity and reduce dropout rates, ultimately accelerating drug development by improving the efficiency of clinical trials.[74]
For example, Genentech employed Snorkel AI to extract key information from over 340,000 CTP eligibility criteria. This effort led to better study designs and more accurate participant inclusion/exclusion criteria. The process helped visualize how different eligibility criteria could impact the demographics of trial participants, allowing for more informed decisions about trials. Snorkel AI identified patterns in clinically relevant characteristics and performed demographic trade-off analyses, enabling Genentech to adjust criteria to enhance trial diversity and success.[75]
Improving diversity in clinical trials is essential for addressing health inequities. According to the FDA, concerningly, 75 percent of clinical trial participants for drugs approved by the FDA in 2020 were white, while only 11 percent were Hispanic, 8 percent were Black, and 6 percent were Asian.[76] This lack of diversity is problematic not only because certain diseases are more prevalent in specific underrepresented groups, but also because different populations may respond differently to therapies. For instance, Albuterol, the most-prescribed bronchodilator inhaler in the world, is less effective for Black children compared with their white counterparts. Since 95 percent of lung disease studies were conducted on individuals of European descent, this genetic difference went undetected for years.[77] Scientists later linked specific genetic variants involved in lung capacity and immune response to the reduced efficacy of Albuterol in Black children.[78] In response to a lack of diversity in clinical trials, the FDA issued draft guidance in 2022 encouraging more inclusive representation.[79] Pharmaceutical companies are increasingly leveraging AI, including tools such as TrialPathfinder and Criteria2Query, to safely recruit more diverse, representative participants, helping to improve trial outcomes for underrepresented populations.[80]
Johnson & Johnson Leveraging AI to Diversify Clinical Trials and Transform Precision Medicine
Data science and AI are transforming how J&J discovers, develops, and delivers new therapies to bring transformative medicines to people around the world. J&J is leveraging AI tools to build and scale capabilities that enhance the design, execution, and diversification of its clinical trials to facilitate drug development. For clinical trial design, J&J has developed an interactive, AI-enabled platform, Clinical Studio, which uses real-world data (RWD)—including EHRs and patient registries—and internal operational data to enable digitization of protocols, providing transparency into clinical cost, protocol complexity, and patient burden and helping researchers develop fit-for-purpose protocols for clinical trials, without impacting the scientific rigor of the study. For example, the platform can be used to define and shape inclusion/exclusion criteria and associated outcomes, which has a direct impact on the cohort of eligible patients, including diversifying the patient pool.
J&J’s internal platform, Trials360.ai, is helping to guide clinical trial site selection as well as engagement and patient recruitment efforts. The tool, coupled with clinical and operational expertise, aims to accelerate clinical development and trial recruitment by meeting patients where they are. With AI algorithms applied to real-world and clinical data, J&J can more accurately predict trial locations with the highest probability of enrolling patients, enabling it to place trial sites in locations where patients are, rather than establishing sites where trials have historically taken place. Findings show that sites ranked in the top 50 percent by its AI models enroll up to three times more patients than do those ranked in the bottom 50 percent. J&J has leveraged AI to review clinical data to identify potential data anomalies and query sites and resolve data discrepancies in an ongoing manner.
Further, across therapeutic areas, J&J is leveraging a data-driven, AI approach to support the advancement of diversity, equity, and inclusion (DEI) in clinical trials—from trial planning to execution—aiming to ensure trial results are generalizable to diverse, real-world populations impacted by the diseases J&J is tackling. In immunology, J&J has set ambitious goals for several trials, and as of late 2023, it had surpassed its year-end diversity goals for those trials, five months ahead of schedule—supported by robust recruitment tactics and AI-supported site selection. In oncology, J&J is leveraging data science, coupled with clinical and operational expertise and community engagement, to help advance DEI in its multiple myeloma (MM) studies through clinical trial site selection and patient recruitment.
A recent meta-analysis of 431 MM studies conducted between 2012 and 2022 across the industry shows that only 4 percent of enrolled patients were Black.[81] Early enrollment data for five ongoing J&J MM trials indicate that a combination of human expertise and AI-driven insights has led twice as many Black patients to consent and enroll in clinical trials in the United States as compared with prior trials.
Optimizing Regulatory Submissions
The regulatory submission phase of clinical trials is both resource intensive and time consuming. Regulatory affairs teams serve as the critical link between pharmaceutical companies and regulatory bodies, working to secure approvals in accordance with current guidelines. These teams compile the necessary data and documents for submission while ensuring that applications meet all regulatory requirements. AI can support this process by reducing the time needed to gather and standardize data, as well as by generating drafts based on templates and guidelines. Furthermore, AI can enhance workflows by providing timely insights, automating tasks such as data monitoring, document management, and adverse event detection as well as ensuring compliance with evolving regulations.[82]
For example, Medidata’s Clinical Data Studio uses AI to monitor clinical trial data in real time, streamlining clinical data management, operations, and safety. The platform automates data reviews, identifies patterns through visualizations, monitors trial site performance and compliance, and mitigates data quality risks by detecting anomalies such as inconsistencies or outliers in patient data. This proactive approach enables clinical researchers to address potential issues early, helping to ensure data integrity throughout the trial and reducing the risk of compromising regulatory compliance. Ultimately, this helps improve trial efficiency and reliability, expediting the regulatory review process.[83]
Another notable example is Veeva Vault RIM (Regulatory Information Management), an AI-powered platform developed by Veeva Systems. The platform offers a comprehensive suite of services designed to manage submissions, ensure compliance, and streamline regulatory processes across regions. With its compliance tracking features, Veeva Vault RIM continuously monitors and updates evolving regulatory guidelines worldwide, enhancing the efficiency of the regulatory submission process.[84] When new guidelines are issued by regulatory bodies such as the FDA or EMA, the platform alerts clinical trial teams, analyzes the changes, and recommends adjustments to trial procedures or documentation in real time to ensure ongoing compliance. Additionally, the platform automates the regulatory submission process by aggregating data from various sources, formatting it according to the latest guidelines, and generating submission-ready documents, which reduces manual effort and the risk of errors.[85]
Enhancing Manufacturing and Supply Chains
AI can also enhance drug manufacturing and strengthen supply chains by optimizing production processes, reducing downtime through predictive maintenance, improving demand forecasting, and streamlining inventory management, ultimately leading to greater efficiency.
AI in Manufacturing
In addition to its role in drug discovery, clinical trials, and regulatory submissions, AI can enhance manufacturing and help strengthen supply chains. It can optimize drug manufacturing by streamlining production processes, improving quality control, and reducing costs. Predictive analytics can identify efficient production pathways, forecast equipment failures, anticipate necessary maintenance, and improve resource allocation. AI can also support the development of continuous manufacturing systems, enabling more flexible, responsive drug production, and AI-driven demand forecasting can help reduce both stockouts and overproduction.[86]
Asimov—Innovating Gene Therapy Design and Manufacturing With AI
Asimov, a Boston-based pioneering bioengineering company, uses AI and synthetic biology to transform the design and production of therapies. Genetic engineering, the process of modifying DNA, holds immense potential for drug development by targeting diseases at their genetic roots. The process is vital for developing biologics, cell and gene therapies such as CAR-T, and precision medicine to tackle complex conditions such as cancer and inherited genetic disorders. Asimov’s AI-driven molecular simulations and computational biology accelerate the traditionally slow, labor-intensive, and trial-and-error genetic engineering process, making drug development faster, cheaper, and more reliable.
Asimov’s AI-driven bioengineering tools apply to both drug design and manufacturing. In drug design, AI generates novel DNA sequences and simulates complex biological processes, expediting the identification of promising drug candidates. In manufacturing, these tools can increase the precision and efficiency of therapeutics production, supporting the creation of complex biologics such as antibodies and viral vectors. This is critical for improving the scalability, reliability, and cost effectiveness of drug manufacturing, especially for targeted cell and gene therapies that require customized production methods.
Asimov’s AAV Edge platform is an example of the company’s innovation in gene therapy manufacturing. The platform optimizes the production of adeno-associated viruses (AAV), a type of virus used in gene therapies. Its AI-designed tissue-specific promoters restrict gene expression to intended tissues, ensuring, for example, that a therapy targeting heart disease is “on” in heart tissue but “off” in liver tissue to reduce toxicity. This is critical because while gene therapies hold significant potential, a key challenge is that they can cause liver toxicity. AAV Edge helps increase the precision, effectiveness, and safety of gene therapies while reducing side effects.[87]
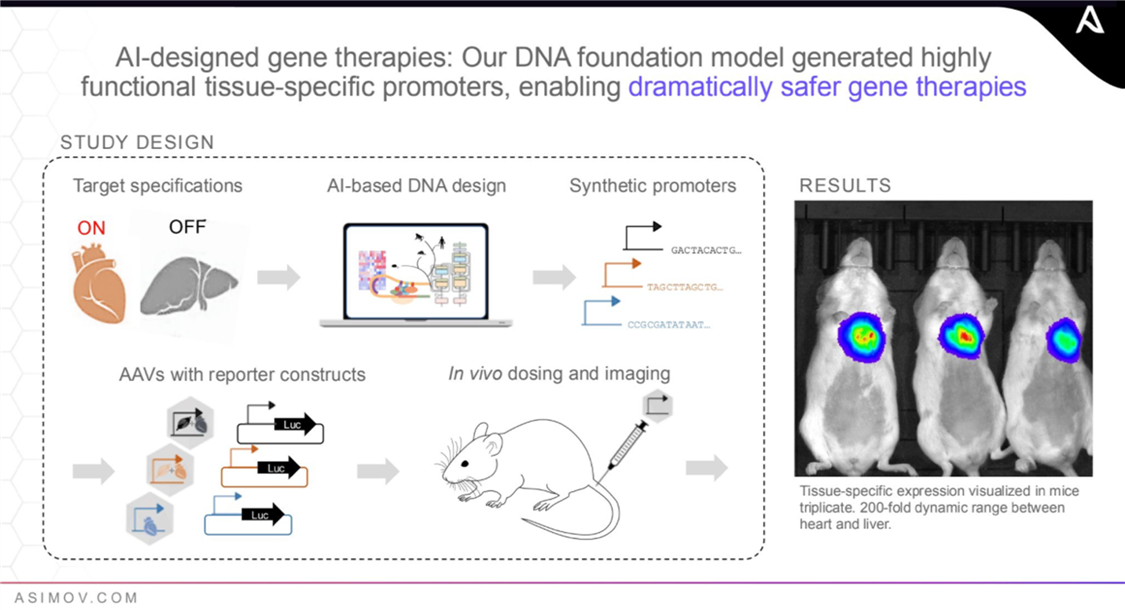
Currently, Asimov focuses on drug manufacturing, particularly drugs enabled by genetic engineering, including antibodies, protein biologics, cell and gene therapies, and RNA vaccines. By employing AI tools, Asimov’s suite of services seeks to improve drug efficiency and accelerate manufacturing timelines, yielding higher, faster, cheaper, and better therapeutic production. Asimov’s future vision is to extend its AI-driven bioengineering solutions to the entire biotech space, including agriculture and food, industrial biotechnology, and environmental biotechnology, to accelerate innovation in these areas by automating traditionally slow, manual processes, increasing precision, and enabling customized solutions.
AI in Supply Chains
Drug shortages are a growing public health concern, with significant financial impacts on health-care systems. In the United States alone, these shortages cost an estimated $230 million a year due to health-care expenses, productivity losses, and adverse patient outcomes. The root causes of such shortages include supply chain management challenges, inadequate business continuity planning, and market dynamics such as fluctuating demand.[88]
The global pharmaceutical supply chain constitutes an elaborate network of manufacturers, suppliers, and distributors spanning multiple countries, rendering it vulnerable to disruptions from natural disasters, transportation delays, and regulatory hurdles. AI can help mitigate these risks by optimizing supply chain processes.[89] It can analyze transportation costs, lead times, and supplier performance, enabling better route planning, cost saving, and improved delivery times. For example, the United States Pharmacopeia’s (USP’s) Medicine Supply Map employs AI to analyze millions of data points worldwide to predict disruptions, allowing timely interventions to prevent shortages.[90]
Overall, AI can drive improvements in manufacturing efficiency, supply chain management, and the scalability of advanced therapies, contributing to faster, more cost-effective drug production and supply chain resilience.
Merck employs an AI system developed by Aera to optimize its supplier network through data analysis and proactive recommendations. This system enables Merck to anticipate supply chain fluctuations, adjust production schedules, and reroute shipments, thereby enhancing efficiency and improving supply chain resilience.[91]
Overall, AI can drive improvements in manufacturing efficiency, supply chain management, and the scalability of advanced therapies, contributing to faster, more cost-effective drug production and supply chain resilience.
Challenges for AI Adoption
AI adoption in drug development faces several significant challenges. Data access and privacy issues present substantial hurdles, as stakeholders—including health-care providers, pharmaceutical companies, researchers, and federal agencies—must collect and share vast amounts of data to develop effective AI models. Improved coordination, both within and across organizations, is crucial to achieve this goal. Additionally, validating AI algorithms is essential to address concerns about the “black box” nature of how AI systems arrive at decisions and to mitigate possible biases, thereby building trust in these technologies. Integrating AI with existing workflows can also be challenging, especially in the face of unclear regulatory pathways. Misaligned incentives, including fears of job displacement, may further slow adoption.[92] However, with proactive strategies and enhanced collaboration between public and private sectors, these challenges can be effectively addressed, paving the way for the successful integration of AI in drug development.
Data Access, Quality, and Privacy
AI can play a crucial role in supporting drug development, but its effectiveness hinges on the availability and quality of training data. Several types of data are essential for AI applications in this area. First, clinical data, including EHRs, provides information on patient medical histories, diagnoses, lab results, imaging data, and treatment plans. Second, genomic data, obtained through sequencing techniques, helps identify genetic variants and provides insights into gene function and regulation, which are vital for understanding disease mechanisms. Third, pharmaceutical data includes information on drug efficacy, adverse reactions, pharmacokinetics, and pharmacodynamics. Fourth, chemical and structural data is important for identifying and optimizing drug candidates from extensive compound libraries. Finally, data from scientific literature—such as research papers, clinical guidelines, clinical trial outcomes, and patents—helps synthesize existing knowledge and identify gaps for new drug development.[93] Together, these different types of data create a comprehensive picture that enhances the ability to identify promising therapeutic targets and drug candidates, target treatments to individual patients, and streamline the drug development process. Importantly, linking genotypes (genetic information found in genomic records) to phenotypes (observable traits found in clinical records) enables researchers to better understand how genetic variations influence disease risk and treatment responses and disease processes. This connection is particularly important for developing targeted therapies and advancing precision medicine. Yet, challenges remain in collecting, sharing, and linking this data to support AI applications.
Two decades ago, the Human Genome Project (HGP) successfully decoded the human genetic code. A core principle that contributed to the success of this international scientific effort was data sharing.[94] HGP leadership established the Bermuda Principles, which committed all project participants to electronically share data and make human genome sequences publicly available in order to advance scientific progress.[95] Since then, advances in sequencing techniques have generated vast amounts of genomic data from millions of individuals, now stored in repositories around the world. The Bermuda Principles, adopted by journals and funding agencies, aim to ensure that published genome study data remains accessible to all, fostering further scientific discoveries.[96]
However, the vast influx of diverse and sensitive data has prompted governments, funding agencies, and research consortia working with them to develop custom databases for managing this information. The existence of incompatible and non-shareable datasets further complicates efforts to link genomic and phenotypic data, which is key for breakthroughs.[97] Uncovering the genetic causes of complex diseases such as cancer and cardiovascular disorders requires pinpointing multiple genetic risk factors across the genome. This is achieved through genome-wide association studies (GWAS), which analyze the genotypes of hundreds of thousands of individuals, both with and without the disease, to identify relevant genetic variations.[98] These studies integrate both genomic and phenotypic data, which often come from EHRs and medical cohort studies—such as the Framingham Heart Study, which tracked multiple generations to investigate cardiovascular risk factors—providing insights into how genetic variations contribute to disease. Yet, data integration problems present challenges for GWAS, as the success of these studies relies on the integration of large-scale genomic and phenotypic datasets.
Since 2005, over 10,700 GWAS have been conducted, generating vast datasets largely stored in controlled-access databases to protect personal information for legal and ethical reasons. Researchers must navigate strict vetting processes to access such data. For example, National Institutes of Health (NIH) grant recipients are required to deposit their GWAS data into an official repository, the Database for Genotypes and Phenotypes (dbGaP), while European researchers are encouraged to use the European Genome-Phenome Archive (EGA) housed at EMBL-EBI. Yet, despite such efforts, the process of accessing such publicly available data remains cumbersome.[99] Moreover, a growing number of countries around the world have initiated large-scale genomic sequencing efforts on their own populations, including America’s All of Us Research Program, Genomas Brasil, the Qatar Genome Program, the Turkish Genome Project, and the Korean Genome Project, among others. Such global genomic sequencing initiatives are crucial for capturing genetic diversity across populations, which can lead to more inclusive and effective biopharmaceutical research. For now, though, how the data from these initiatives will be shared and integrated into existing workstreams and consortia remains unclear, diluting the potential of such large-scale datasets.
Initiatives such as the Global Alliance for Genomics and Health (GA4GH) work to create technical standards to link disparate genomic databases globally.[100] Additionally, the GWAS Catalog, an open-access collaboration between EMBL-EBI and the National Human Genome Research Institute (NHGRI), is working to standardize and centralize GWAS data to support both a deeper understanding of disease mechanisms and the discovery of new therapeutic targets and causal variants.[101] Other efforts, such as the Human Cell Atlas (HCA), a global consortium creating publicly accessible, detailed reference maps of human cells, further seek to support AI-enabled drug development and advance our understanding of health and disease.[102]
Electronic Health Records
In addition to genomic data, clinical data—often stored in EHRs—provides crucial details about patients’ medical histories and plays a vital role in deepening our understanding of disease mechanisms. Together, these two types of data are essential for uncovering how genetic variations (genotypes) influence observable traits (phenotypes) and disease outcomes, forming the foundation for building effective AI models for drug development.
The push for electronic health information exchange began in 2009 with the signing of the American Recovery and Reinvestment Act (ARRA), which established the Health Information Technology for Economic and Clinical Health (HITECH) Act. This provision promoted the digitization of patient records to improve health-care delivery, allocating over $35 billion to support the adoption of EHRs by hospitals and clinics.[103] Today, EHR adoption is widespread, with 96 percent of U.S. hospitals and almost 80 percent of office-based physicians using them.[104] Some of the largest EHR systems include Epic Systems, Oracle Cerner, and MEDITECH. Epic holds the largest market share at 37.7 percent, followed by Oracle Cerner at 21.7 percent, and MEDITECH at 13.2 percent.[105] Epic, the leading provider in the United States, is used by many prominent hospital systems and academic medical centers, including the Cleveland Clinic, the Mayo Clinic, and Johns Hopkins Medicine.
Beyond the issues posed by privacy concerns, the presence of hundreds of EHR systems across the United States, each with distinct clinical terminologies and technical standards, complicates interoperability. True interoperability requires not just the exchange but also the use of standardized data, a long-standing issue in the U.S. health-care system that continues to limit electronic data sharing. Moreover, achieving interoperability depends on collaboration between many stakeholders, including patients, providers, software vendors, legislators, and health information technology (IT) professionals. Yet, the health-care system remains fragmented, with data often treated as a commodity for competitive advantage rather than a shared resource for improving patient care.[106]
Recently, the FDA pushed for an increased use of RWD—including EHRs, billing data, and administrative claims—in drug development. This data, rich in patient details including disease status, treatments, procedures, and outcomes, could enhance drug development. With the growing EHR adoption across the United States, the FDA has issued guidance on using EHR data in clinical research and regulatory submissions.[107] However, integrating RWD with AI presents several challenges. EHRs are often inconsistently documented by clinicians, making it difficult to extract data, such as treatment outcomes, uniformly. Issues including missing data and selection bias further complicate the analysis. Additionally, the lack of standardization and harmonization across different data sources hinders both replication and reproducibility.[108]
Despite these challenges, the creation of large research networks such as the national Patient-Centered Clinical Research Network (PCORnet), the Observational Health Data Sciences and Information (OHDSI) consortium, and the Clinical and Translational Science Award Accrual to Clinical Trials (CTSA ACT) network have improved data sharing. These networks span multiple sites around the world, employing standardized data infrastructure, and provide access to a diverse patient population, enabling large-scale studies to explore factors influencing health and disease.[109]
EHR-driven genomic research is valuable, as an integrated approach can augment GWAS by replicating the studies and extending conventional GWAS findings to underrepresented populations.
In 2013, the National Science Foundation (NSF) convened a workshop to identify challenges and set a research agenda to achieve a national-scale Learning Health System (LHS). An LHS is an infrastructure that enables rapid data sharing and knowledge generation to inform health-care decisions, ultimately improving health outcomes. The agenda emphasized the need for systematic integration of data, evidence, and practice, highlighting the importance of standardizing data stored in EHRs. By facilitating access to comprehensive patient data, EHRs enable health-care providers to engage in continuous learning. The agenda noted that, to maximize the potential of EHRs in fostering a collaborative, well-functioning LHS, establishing standardized data-sharing protocols and ensuring interoperability are essential for adaptation and innovation in response to emerging health-care challenges.[110]
Linking EHRs with other data sources, including genomic data, can further enable the study of drug-phenotype and drug-gene interactions. Researchers from the Vanderbilt Electronic Systems for Pharmacogenomic Assessment (VESPA) project have shown that EHR-based biobanks—repositories of human biological materials—provide cost-effective tools for biomedical discoveries, as they allow for the reuse of biological samples across multiple studies and enhance research efficiency.[111] Such EHR-driven genomic research (EDGR) is valuable, as an integrated approach can augment GWAS by replicating the studies and extending conventional GWAS findings to underrepresented populations. This is because EDGR cohorts reflect the populations cared for in clinical settings, which are much more diverse than those typically represented in genomic research.[112] EHR-linked genomic studies can also enable phenome-wide association studies (PheWAS), which analyze the effect of genetic variants across multiple diseases and traits captured in EHRs. A major obstacle, however, remains the need for the development and adoption of international standardized consent models from patients for the use of their clinical data in biomedical research, as consent regimens vary significantly across regions.[113]
Lucia Savage, chief privacy and regulatory officer at Omada Health and former chief privacy officer at the U.S. Department of Health and Human Services (HHS) Office of the National Coordinator for Health IT, has highlighted that regulations governing the use of data collected through traditional health-care processes vary widely across countries. In the United States, there is more leeway for using patient data in tertiary research applications, such as developing AI models for drug development, as the Health Insurance Portability and Accountability Act (HIPAA) allows the use of patients’ health data for research without explicit consent when that data is de-identified. In contrast, many other countries, including those governed by the EU’s General Data Protection Regulation (GDPR), impose stricter restrictions on such uses and the international transfers of personal data. These differences in legal frameworks make cross-border data sharing and access more challenging. Therefore, it is crucial to consider the regulatory environment in which the data was collected and explore possibilities for interoperability to navigate such challenges.[114]
Gilead Using AI to Identify Underdiagnosed Hepatitis C Individuals[115]
Gilead is leveraging AI to identify underdiagnosed individuals, particularly for diseases such as HIV and hepatitis C virus (HCV), where traditional screening methods are expensive and burdensome for patients. In diagnosis, there is a trade-off between privacy and data usability, as often, the higher the privacy standard, the less usable the data. Hence, striking a balance between privacy and usability is key. Too much privacy can make it difficult to identify patients, particularly from underserved communities. Greater data access, sharing, and interoperability, supported by privacy-enhancing techniques, could help with more accurate diagnoses.
Gilead’s machine learning algorithm, trained on ambulatory electronic medical records (EMRs), aims to predict initial HCV diagnoses and identify undiagnosed HCV patients, prioritizing them for screening. The EMRs used to train the algorithm include data on age, gender, HCV-related predictors such as birth cohort, opioid usage, laboratory test results, diagnosis codes, treatments, data on social determinants of health, chronic health conditions, and other variables.
HCV, one of the most common blood-borne viruses and a leading cause of liver-related illness in the United States, is the target of a World Health Organization (WHO) initiative to eradicate it as a public health threat by 2030. The National Academies of Science, Engineering, and Medicine (NASEM) have highlighted improved detection of undiagnosed HCV cases as central to eliminating the virus. Universal one-time screening is recommended in the United States, but it is difficult to implement in practice, and screening rates remain low. Gilead’s AI approach, trained on a large EMR dataset, requires fewer patients to be screened compared with traditional methods while improving precision. The AI model can prioritize patients for HCV screening, and has the potential to make resource allocation more efficient, reduce clinician workload, prevent disease progression, and lower health-care costs.
Integrating AI into EMR systems and clinical workflows shows significant potential for accelerating HCV elimination efforts. Effective targeting could improve diagnosis rates, reduce morbidity and mortality through earlier detection, and identify patients often overlooked by risk-based screening. The AI algorithm generates continuous risk scores, allowing for a more nuanced triage process and targeted screening—patients with higher scores calculated from their EMR data can be prioritized for screening, diagnosis, and linkage to care. This data-based approach can help identify harder-to-find patients in a way that does not stigmatize individuals. It could also improve the allocation of finite health-care resources and the return on investment of screening programs, as well as the rates of HCV diagnoses, treatment, and transmission.
Gilead’s AI use case demonstrates how machine learning and EMRs, when combined, present new opportunities to improve population health management and achieve better clinical outcomes. Moreover, it supports public policies that promote EMR adoption and interoperability, which could play a critical role in identifying underdiagnosed individuals, particularly within underserved communities. By enhancing data sharing and integration, these policies could not only streamline clinical workflows, but also help reduce health disparities, supporting efforts to improve health equity and ensure timely, effective care for all patients.
Beyond RWD, there is growing interest in synthetic data—artificially generated by computer algorithms to simulate the statistical properties of RWD—as a scalable, cost-effective, and privacy-preserving alternative for training AI models. However, challenges remain: Synthetic data may not fully capture the complexity and variability of real clinical populations, such as diverse demographics and the intricate biological and clinical interactions that influence treatment responses and side effects. There are also questions about how well synthetic data reflects the evolving nature of diseases, treatment protocols, and patient populations. The use of synthetic data in drug development remains an emerging area of research.[116]
Pharmaceutical Data
Pharmaceutical data, alongside genomic and clinical data, plays a key role in biopharmaceutical innovation. While biopharmaceutical companies often prefer to keep their data private for competitive reasons, collaboration between companies and research institutions could significantly speed up drug development. Public-private partnerships (PPPs) that bring together academia, industry, and government offer a path forward, particularly in precompetitive research, where the risks are lower. One notable example is the Innovative Health Initiative (IHI), the largest biomedical PPP in the world, launched in 2008 by the European Union and the European Federation of Pharmaceutical Industries and Associations (EFPIA) to accelerate the development of next-generation therapies.
By encouraging the sharing of data among academia, industry, and government, particularly in precompetitive research, PPPs are producing valuable training data for AI-enabled drug development.
Other examples include the Alzheimer’s Disease Neuroimaging Initiative (ADNI), launched by the NIH’s National Institute on Aging (NIA) to advance Alzheimer’s research; Project Data Sphere, launched by the CEO Roundtable on Cancer to accelerate drug development by facilitating access to de-identified oncology clinical trial data; Open Targets, which uses biological data to identify and validate therapeutic targets; and the Structural Genomics Consortium, focused on advancing knowledge of human protein structures.[117] By encouraging the sharing of data among academia, industry, and government, particularly in precompetitive research, PPPs are producing valuable training data for AI-enabled drug development.
Beyond PPPs, alternative payment models can provide incentives for pharmaceutical companies to share data. Data monetization allows companies to profit from their data by providing access to anonymized datasets on a subscription basis. For example, Flatiron Health, acquired by Roche, operates under this model by aggregating and analyzing de-identified oncology RWD to offer insights that enhance cancer care and accelerate drug development. This model treats data as a product, creating a continuous revenue stream for companies.[118]
Role of Privacy-Enhancing Technologies
When sharing data, a key consideration is ensuring that it can be done in a secure, privacy-enhancing manner, given the confidential and sensitive nature of the data involved in drug development. Traditional methods of sharing data between different entities involve sending data to third parties, such as by creating copies of data for each entity or aggregating data in a single repository that all entities can access. Entities can maintain privacy of shared data through access controls, oversight, and legal mechanisms. However, newer decentralized methods for data sharing allow multiple entities to collaborate on AI model training without sharing raw internal data. Decentralized data sharing keeps data distributed across multiple locations, eliminating the need to transfer or aggregate it in a central repository. This method is particularly valuable in privacy-sensitive settings, as it enables collaboration while safeguarding sensitive information. In drug development, decentralized approaches enable different research institutions and companies to share data, supporting the development of safer, more effective therapies.[119]
PETs can support the training of AI algorithms on vast biological, chemical, and clinical datasets, accelerating AI-enabled drug development.
Key aspects of decentralized data sharing include a strong emphasis on data privacy, collaboration, and security. In this approach, each party retains control over its data, lowering the risk of leaks from third parties and minimizing the threat of a single point of failure that could compromise the entire dataset. Decentralized approaches are often supported by privacy-enhancing technologies (PETs), such as secure multiparty computation (SPMC), federated learning (FL), fully homomorphic encryption (FHE), and differential privacy (DP), which enable privacy-enhancing access to and analysis of diverse data sources.[120] PETs can support the training of AI algorithms on vast biological, chemical, and clinical datasets, accelerating AI-enabled drug development.[121]
Secure Multiparty Computation
SPMC is a cryptographic method that enables multiple participants to collaborate on computations using private data without revealing it to each other, allowing teams to work together with internal data while maintaining its confidentiality. SPMC ensures that only the final results of a computation are revealed to participants without disclosing any intermediate information from a joint analysis, thereby providing a higher level of security.[122]
In drug discovery, SPMC has several important applications. For instance, it can be used to predict interactions between therapeutic targets and drugs based on genomic and chemical data. This is a critical step in developing promising drugs, and large datasets are required to train AI models to generate accurate predictions. Public datasets, including ChEMBL—a database of bioactivity data for drug-like compounds—often rely on contributions from academic institutions, pharmaceutical companies, and collaborative projects and help train such models on vast data. But they are only one piece of the puzzle. The integration of other sensitive data, including EHRs, could further enhance algorithms. Methods such as SMPC can help, and feasible SMPC protocols have been deployed for GWAS and drug-target interaction prediction problems. However, efforts to develop comprehensive SMPC protocols for AI-enabled drug discovery are still a work in progress.[123]
Consider how SMPC can be useful in drug discovery. Pharmaceutical companies, even competitors, benefit from collaboration but must protect their proprietary data. For example, Company A, which specializes in high-throughput screening of small molecules, and Company B, which has comprehensive datasets of biological targets, can use SMPC to securely combine their datasets to discover compounds that interact with certain biological targets. In this manner, they can jointly identify potential drug candidates for further exploration, all while safeguarding each company’s proprietary data and enhancing their research.[124]
Federated Learning
FL emphasizes collaborative training of AI models while keeping the training data decentralized and local to each participant. In this approach, each participant trains the AI model on their own data and shares only the model parameters—rather than raw data—with a central server, which aggregates these parameters to enhance the global AI model. The refined model is then returned to the participants for further local training, enabling collaborative model development without exchanging sensitive data. Unlike SMPC, which allows multiple parties to jointly perform computations on private data, FL centers on the collaborative building of models. For instance, hospitals could use FL to collaboratively train a model that predicts patient outcomes based on their EHRs without exchanging any actual patient data.[125]
In drug discovery, FL is particularly dynamic and offers numerous applications. A notable example is IHI’s MELLODDY (MachinE Learning Ledger Orchestration for Drug DiscoverY) project, which embodies a blend of cooperation and competition. This initiative emerged from a joint call by 10 of the world’s largest pharmaceutical companies aimed at developing predictive AI models for drug discovery while safeguarding data privacy. The collective dataset for this project will encompass over 10 million small molecules and more than 1 billion activity labels measured in biological assays, making it one of the largest FL-based efforts in the field.[126]
Fully Homomorphic Encryption
FHE is another powerful technique that enables computations on encrypted data without requiring decryption, ensuring that data remains secure.[127] In 2017, experts from industry, government, and academia established the Homomorphic Encryption Standardization Consortium, which developed a standard in 2018 outlining security requirements for FHE applications.[128]
In drug discovery, FHE presents a promising solution to privacy concerns by allowing scientists to perform computations on encrypted data, thus protecting sensitive information such as the structures of new drug compounds and genomic data.[129] While algorithmic improvements have already enhanced the efficiency of FHE, widespread implementation still faces challenges, including high computational demands and integration with existing data workflows. Advances in hardware acceleration and optimized algorithms could, however, enhance its use in privacy-enhancing decentralized AI model building.[130]
Differential Privacy
DP, introduced by Cynthia Dwork in 2006, is a mathematical concept that enables data sharing while preserving individual privacy. The core principle of DP is that the outcome of a computation should remain nearly the same whether a single data point is included or excluded. This is achieved by adding calibrated noise to the results, effectively hiding individual contributions while maintaining the overall accuracy of the analysis. Although DP results are approximate and may vary with repeated analyses, sensibly calibrated noise enables AI models to balance privacy and utility. Still, DP requires careful tuning of noise levels to ensure that data utility is not compromised.[131]
DP has proven particularly useful in applications such as drug sensitivity prediction. For example, a 2022 study shows that combining DP with deep learning, a technique known as differentially private deep learning, can effectively predict breast cancer status, cancer type, and drug sensitivity using genomic data, all while preserving individual privacy.[132]
Beyond the importance of advancing the development of PETs—including SPMC, FL, FHE, and DP—to enable secure data sharing in AI-enabled drug development, it is crucial to make these tools accessible and user friendly for a wide range of scientists working in biopharmaceutical innovation. Organizations at the forefront of PET development include U.S.-based OpenDP, Duality Technologies, and Actuate, as well as the United Kingdom’s OpenMined.[133] For example, Duality Technologies has collaborated with leading research institutions such as the Dana-Farber Cancer Institute to enhance oncology outcomes.[134]
Algorithm Validation and Bias Mitigation
As AI becomes increasingly integral to drug development, attributes such as model accuracy, reliability, transparency, and interpretability are important for creating effective therapies. However, the complexity of neural networks underlying AI often causes these models to function as black boxes. To tackle this challenge, eXplainable AI (XAI) is emerging as a branch of AI focused on developing models that are clearer and more interpretable.[135] This enables researchers to identify and correct potential errors, such as inaccurate predictions due to flaws in the training data.[136] Furthermore, drug development is an increasingly multidisciplinary process involving chemists, biologists, clinicians, and data scientists, and XAI helps make AI-driven insights more accessible to non-AI experts, fostering collaboration. The effective integration of human expertise with AI insights is essential for accelerating the development of safe and effective therapies.[137]
The National Institute of Standards and Technology (NIST) defines trustworthy AI as “valid and reliable, safe, secure and resilient, accountable and transparent, explainable and interpretable, privacy-enhanced, and fair with harmful bias managed.”[138] Organizations such as NSF’s Institute for Trustworthy AI in Law & Society (TRAILS) at the University of Maryland and the AI Now Institute are working to promote these principles for the use of AI across different industries, including biopharmaceuticals.[139]
An increasingly important reason to validate AI is to identify and mitigate potential biases in the models. The FDA defines algorithmic bias as “the systematic deviation in model predictions or outcomes for certain data points or groups compared to others.”[140] Bias can emerge at different stages of the drug development process—from biases in clinical research data used to identify new therapeutic targets and drug candidates, which may disadvantage underrepresented populations, to biases in manufacturing data that can undermine model performance and limit generalizability in the production of therapies.
Without human oversight, AI systems can unintentionally learn and perpetuate biases present in their training data. In drug development, such biases may lead to inequitable outcomes, such as underrepresenting certain patient groups, which can skew predictions about drug efficacy or safety. This could result in therapies that are less effective, or even harmful, for these populations. Validating AI models to detect and correct biases ensures that these technologies support fairness and inclusivity in drug development, producing safer, more effective therapies for all patients.
To promote equity in AI-supported drug development, it is important to use diverse, representative datasets. Yet, a 2009 study revealed that 96 percent of GWAS participants were of European descent, a figure that decreased to 78 percent by 2018.[141] The lack of diversity in genomics research could introduce bias in AI algorithms and exacerbate health-care disparities if left unaddressed by both failing to account for differences in drug response among diverse groups and neglecting diseases prevalent in underrepresented populations.[142] For example, as previously mentioned, Albuterol, the most-prescribed bronchodilator inhaler, is less effective in Black children due to genetic differences that had gone unnoticed for years, as 95 percent of lung disease clinical trial participants were of European descent. Identifying the specific genetic variants that contribute to differences in Albuterol response has both enhanced scientists’ understanding of drug efficacy and highlighted the value of inclusive research.[143]
To address such biases, it is essential to use more diverse datasets that represent various demographic and clinical attributes and employ techniques such as data reweighting to balance underrepresented groups, cross-population model validation, and regular audits. Moreover, expanding genetic sequencing efforts and integrating clinical data from underrepresented groups through EHRs could help, because EHR data better reflects the diverse populations seen in real-world clinical settings.[144]
Beyond the potential consequences of algorithmic bias in clinical research, data biases can also arise in drug manufacturing, harming model performance and generalizability. For example, variability between production batches (e.g., differences in raw materials or slight equipment changes) can introduce biases if AI models are trained disproportionately on data from a particular batch, leading to inaccurate predictions and process adjustments that do not generalize across batches. Mitigating algorithmic bias throughout drug development, from clinical research to manufacturing, is therefore essential.
Efforts to Reduce Algorithmic Bias
Several efforts are underway to improve the representativeness of data used in biomedical research. One notable example is the NIH’s All of Us research program, launched in 2018. This large-scale genetic sequencing initiative aims to advance health equity by gathering data from a diverse population. With over $3.1 billion in funding, it seeks to create the world’s most diverse genetic dataset for biomedical research. Currently, 80 percent of All of Us participants come from groups historically underrepresented in biomedical research, spanning different races, ethnicities, ages, geographies, access to health care, and disability status. The data includes survey responses, EHRs, genome sequences, and activity data from devices such as Fitbit. Researchers can access this information through a centralized cloud-based workbench. Since its first data release in 2020, over 200 peer-reviewed articles have been published based on this data.[145]
All of Us fills a critical gap in genomics research by providing the enhanced diversity that is largely absent from most major biobank resources. For example, the UK Biobank, the world’s largest whole-genome dataset, has released about half a million genomes, with approximately 88 percent coming from white individuals.[146] A study on type 2 diabetes illustrates the strength of having a diverse dataset. With data from over 2.5 million individuals—also drawing from All of Us—including nearly 40 percent from non-European ancestries, the researchers identified 611 genetic markers that could influence the development and progression of diabetes, 145 of which were previously unknown. These discoveries hold the potential to guide more precise, genetically informed diabetes care. Furthermore, population-specific differences in genetic variants linked to drug metabolism underscore that certain drugs may be safer or more effective for specific groups. Such findings are crucial in ensuring that AI-enabled drug development is equitable and reflective of diverse populations.[147]
All of Us fills a critical gap in genomics research by providing the enhanced diversity that is largely absent from most major biobank resources.
Integration With Workflows
AI has the potential to reshape production processes in biopharmaceutical innovation, quickly scanning vast datasets to uncover correlations and make predictions. But AI should be viewed as a complement to, rather than a substitute for, human scientists. While AI excels at detecting patterns, it cannot discern causal relationships—such as whether a protein implicated in a disease actually causes the disease or is merely associated with it. Human expertise remains crucial for designing effective drugs, as scientists uncover and map such critical cause-and-effect relationships, in which AI models can then ground their predictions.[148] This can also help alleviate potential concerns about job displacement.[149] By repackaging a job’s tasks into distinct categories—some suited for humans, and others for AI—we can clarify their respective, complementary roles in drug development.[150]
Effective integration of AI into drug development workflows is crucial for leveraging both AI’s predictive power and human judgment to accelerate the development of safe and effective therapies.[151] This integration involves substantial up-front costs for staff training and workflow system adjustments. Implementing these changes requires coordinated efforts, complementary assets, financial resources, management support, and long-return investment perspective.[152]
Policy Recommendations
Supporting the adoption of AI in drug development requires a comprehensive approach that combines public policies to address a number of technical, operational, and regulatory challenges while fostering innovation. The following are several recommendations for policies that could promote effective AI integration in drug development.
Support for Privacy-Enhancing Data Sharing and Access
A 2019 Information Technology and Innovation Foundation (ITIF) report argues in support of the development of a National Health Research Data Exchange in the United States, which could increase the availability of data to support data-driven drug development.[153] One prominent initiative in this direction, which seeks to facilitate the secure, effective exchange of health data and advance interoperability on a national level, is the Trusted Exchange Framework and Common Agreement (TEFCA). TEFCA was introduced as part of the 21st Century Cures Act of 2016 and implemented by HHS. Formally launched in early 2022, TEFCA went live at the end of 2023.[154] It aims to create a nationwide network for the secure exchange of health data, enabling interoperability and sharing of EHRs among health-care providers, patients, and public health agencies.[155] Seven organizations are currently designated as Qualified Health Information Networks (QHINs) under TEFCA, facilitating data sharing across various stakeholders, including Epic Nexus, eHealth Exchange, and Health Gorilla.[156]
Additional public policies supporting EHR adoption, such as proposed legislation aimed at enhancing the use of EHRs in behavioral health services, could further help.[157] Policies that promote the standardization of data formats across the biopharmaceutical ecosystem, making them less fragmented and disparate, would also enable AI models to share and access diverse datasets, enhancing robustness and effectiveness.[158] Moreover, EHR systems require a deeper conceptual framework that can systematically capture the complexity of clinical mental models and integrate additional data, such as patient-reported outcomes, to improve AI model training.
PETs could further support and encourage the sharing of different sources of data. While significant advances have been made in PETs, more progress is needed in their development, implementation, and adoption. Recent policy efforts aim to bolster these advancements. In October 2023, the White House issued the Executive Order Advancing a Vision for Privacy-Enhancing Technologies spearheaded by the Office of Science and Technology Policy (OSTP).[159] This order calls for the design, development, and deployment of PETs. Subsequently, in April 2024, the Privacy Enhancing Technology Research Act (H.R.4755) passed the House and as of this writing is under consideration in the Senate.[160] This legislation would mandate NSF to support research into PETs and directs OSTP to coordinate efforts with other federal agencies to accelerate their development, deployment, and adoption. Key organizations driving PET innovation include U.S.-based OpenDP, Duality Technologies, Actuate, and U.K.-based OpenMined.[161] Policies that support PET advances can foster increased use of sensitive data in biomedical research and thus accelerate AI-enabled drug development.[162]
Further, governments could support the establishment of data banks—large-scale repositories that collect clinical, genetic, and pharmaceutical data—to enhance the training of AI models. A notable example is the NIH’s All of Us program, which, by creating a comprehensive and diverse data resource, can serve as training data for AI-enabled biopharmaceutical research. Another example is the Cancer Moonshot Initiative, which fosters collaboration and data sharing across the health care and pharmaceutical sectors to advance cancer treatment.[163]
In addition to privacy-enhancing data sharing approaches, other strategies could further encourage cooperation across the biopharmaceutical ecosystem. One promising approach is the establishment of PPPs to foster collaboration among industry, academia, and government stakeholders. Such PPPs can align the interests of public institutions and private companies by addressing key concerns such as IP rights and competitive advantages. While biopharmaceutical companies often keep their datasets confidential to maintain a competitive advantage, fostering collaboration between companies and research institutions could accelerate AI-enabled drug development. This collaborative approach could lead to safer, more transparent, more reproducible innovation processes, while also reducing redundancy and associated risks.[164]
A number of PPPs have emerged to focus on precompetitive research, a phase in which data sharing does not compromise a company’s competitive edge. In this setting, companies can benefit from shared research costs, access to larger and more diverse datasets, and streamlined processes. This model has been successfully implemented in different regions. For example, IHI, the world’s largest biomedical PPP, aims to enhance the EU’s competitive position in pharmaceutical research and accelerate the development of next-generation therapies through initiatives such as its aforementioned federated learning platform MELLODDY.
Policies that support PET advances can foster increased use of sensitive data in biomedical research and thus accelerate AI-enabled drug development.
Another example is the ADNI, launched in 2004 the NIH’s National Institute on Aging (NIA). It remains the largest PPP in Alzheimer’s research, convening leading research centers across the United States, NIA, pharmaceutical companies, and foundations to advance Alzheimer’s disease research. ADNI’s publicly available database, hosted by the Laboratory of Neuroimaging, has also made significant contributions to enhancing the understanding of complex diseases beyond Alzheimer’s, fostering broader scientific discoveries.
As mentioned, another prominent PPP is Open Targets, a collaboration between research institutions such as the EMBL-EBI and the Wellcome Sanger Institute, as well as pharmaceutical companies such as Pfizer, Genentech, and GSK. Its goal is to use biological data to identify and validate therapeutic targets for drug discovery, making data publicly available to accelerate progress.[165]
The Structural Genomics Consortium (SGC) is a PPP that unites academic researchers and pharmaceutical companies such as Johnson & Johnson, Pfizer, and Genentech. It focuses on open-access research aimed at understanding the structure and function of human proteins and generates publicly available data such as protein structures and chemical probes.[166] Building on SGC’s open-science model, a new initiative called Conscience was launched in 2023, in partnership with SGC and supported by a $49 million investment from the government of Canada. Conscience brings together academics, researchers, industry, and community leaders to address market failures in drug development, particularly in areas such as neurodegenerative diseases, pandemic preparedness, rare childhood diseases, and antimicrobial-resistant bacteria. Conscience’s approach relies on two key pillars: AI and open science. By fostering collaboration among companies and researchers, Conscience aims to advance best practices in AI, build tools to accelerate drug discovery, and create benchmarks for shared learning. Its open-science model encourages the exchange of data and discoveries between organizations, reducing duplication, inefficiencies, and costs. Together, these initiatives fuel an entire ecosystem approach to innovation that bridges the gap between industry, academia, government, and nonprofit organizations to accelerate AI-enabled drug development.[167]
These initiatives highlight how PPPs can encourage data sharing in precompetitive research, support the training of AI models, and therefore accelerate AI-enabled drug development when structured effectively.[168] Collaborations between academia, industry, and government can establish secure, privacy-enhancing frameworks for data sharing and analysis, benefiting multiple stakeholders across the biopharmaceutical ecosystem and driving innovation while promoting principles such as reproducibility, transparency, and explainability and reducing risks and redundancies.
Public Funding
Governments could significantly advance research in AI-enabled drug development by increasing funding for key initiatives. For example, NIH’s Bridge2AI Program seeks to accelerate AI adoption in biomedical research through interdisciplinary collaborations and large-scale data creation.[169] Further, the NIH Small Business Innovation Research (SBIR) and Small Business Technology Transfer (STTR) programs provide seed funding to small businesses, including those developing AI solutions for drug development, while NSF’s Directorate for Technology, Innovation, and Partnerships has supported AI-enabled platforms to speed up drug discovery.[170] The Advanced Research Projects Agency for Health (ARPA-H) also leads several initiatives to enhance AI-enabled drug discovery.[171]
Public funding—as a precursor and complement to biopharmaceutical company investments—is essential for several reasons. It supports foundational research, such as AI studies on new drug development algorithms and basic science applications of AI that may not yield immediate commercial returns but can pave the way to major breakthroughs and further incentivize private sector investment. By funding research without direct commercial application or with high uncertainty, public funding can foster innovation that might otherwise be missed. Moreover, public funding can help increase efficiencies, de-risk early-stage innovation, and ensure that AI tools address public health needs, enhancing health equity. This is crucial for tackling unmet medical needs in areas such as antimicrobial/antibacterial research, where private investment may be insufficient.
By funding research without direct commercial application or with high uncertainty, public funding can foster innovation that might otherwise be missed.
For example, ARPA-H’s mission to address large-scale health challenges mirrors past efforts such as the Defense Advanced Research Projects Agency’s (DARPA’s) work to develop the early infrastructure for the Internet and Global Positioning System (GPS) technology. These past successes show how public funding can drive transformative advances by supporting high-risk innovations that can later be commercialized. Public funding also encourages open science and collaboration, resulting in the creation of public goods such as large datasets, tools, and infrastructure that are available to all agents in the drug development ecosystem. Finally, public funding can address ethical and privacy concerns by ensuring that AI development aligns with guidelines such as fairness, transparency, and the mitigation of bias to support equitable AI-enabled drug development.
Governments can also promote collaboration between academia and the private sector to advance AI research and applications in drug development. For example, the Accelerating Medicines Partnership program, a PPP launched between NIH, the FDA, biopharmaceutical companies, and nonprofit organizations in 2014, could be expanded to support AI-enabled drug discovery.[172] Finally, offering incentives for companies investing in AI-based drug development could lower financial barriers to adoption, encouraging the creation of AI tools and their integration into drug development pipelines.
Education and Workforce Development
Governments should also invest in AI and data science training to prepare the next generation of scientists for AI-supported drug development, including updating medical and pharmaceutical curricula to reflect advances in AI. The future of drug development depends on an effective partnership between human scientists and AI technologies, wherein scientists leverage their causal inference skills to convert AI predictions into effective therapies. This collaboration not only enhances the drug development process, but also ensures that therapeutic innovations are grounded in robust scientific understanding. To ensure effective human-AI collaboration, policies must encourage the integration of AI into biomedical research, support cross-disciplinary collaboration, and establish best practices.[173]
Agencies such as NSF can support research on the complexities of human-AI interactions, while the Department of Labor can help prepare workers for an AI-intensive job market through training programs focused on high-level problem-solving and inference tasks. Embracing the strengths of both human creativity and AI can help drive biopharmaceutical innovation. Programs such as NSF’s AI Research Institutes exemplify this approach, advancing AI research and training scientists, including in biomedical fields.[174] Similarly, NIH’s Big Data to Knowledge (BD2K) initiative provides interdisciplinary training to equip researchers with AI and biomedical data science skills to accelerate innovation.[175]
The future of drug development depends on an effective partnership between human scientists and AI technologies, wherein scientists leverage their causal inference skills to convert AI predictions into effective therapies.
Developing an AI talent pipeline is key. Policymakers should promote AI-related education at all levels, ensuring courses count toward graduation requirements, expanding access to STEM-focused charter schools and incentivizing computer science in higher education. As AI reshapes drug development and alters traditional roles, reskilling workers in both the public and private sectors could help ensure the smooth integration of AI into existing workflows.[176] Additionally, policymakers could establish new immigration channels for AI professionals to attract and retain talent in the field.[177] The October 2023 White House Executive Order on the Safe, Secure, and Trustworthy Development and Use of Artificial Intelligence acknowledged the importance of foreign AI expertise and advocated for policy reforms to modernize immigration pathways for AI experts.[178] Such measures could help the United States maintain its leadership in a highly competitive global market for AI expertise.
Regulatory Guidelines
Pharmaceutical regulatory agencies have seen a surge in AI-enabled drug applications and are actively working to understand the regulatory implications of these tools. In 2021 alone, the FDA received over 100 such submissions for AI applications ranging from drug discovery and clinical research to post-market surveillance and advanced manufacturing.[179]
In September 2024, following months of consultations with developers, academics, and regulators on a prior draft, EMA issued a reflection paper on AI in drug development.[180] The paper encourages a risk-based approach for developing, deploying, and monitoring such tools, with risk depending on the specific AI use case. For example, AI applications in drug discovery are typically considered low risk, since drug candidates proposed with AI assistance would still undergo the usual rigorous clinical trial testing. In nonclinical development, AI models that replace or reduce animal testing must adhere to Good Laboratory Practice (GLP). In clinical trials, regulatory risks are lower in early-stage trials (e.g., AI used for patient selection and recruitment) but increase in later stages. In manufacturing, AI uses in process design, optimization, scale-up, and quality control should follow the quality risk management principles to ensure patient safety and product quality.[181]
Policies that clarify and simplify the review process for the use of AI/ML in drug development—and adopt a risk-based approach, recognizing that some applications, such as in drug discovery, are lower risk than others—could encourage the wider adoption of these tools.
In 2023, the FDA released a discussion paper, “Using Artificial Intelligence and Machine Learning in the Development of Drug and Biological Products,” which outlines different use cases of AI/ML in drug development and emphasizes the adoption of a risk-based approach to evaluating such tools, based on the risk to the patient of the particular AI application.[182] The FDA has solicited feedback from stakeholders on its paper, and aims to finalize the guidance by late 2024, intending to safely expand the adoption of AI/ML tools in drug development.[183]
Policies that clarify and simplify the review process for the use of AI/ML in drug development— and adopt a risk-based approach, recognizing that some applications, such as in drug discovery, are lower risk than others—could encourage the wider adoption of these tools. To spur their adoption globally, regulators from different countries could also coordinate to create harmonized standards and approval processes for AI-supported drug development, benefiting both regulatory agencies and pharmaceutical companies operating in multiple countries.
Moreover, the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use, which works to develop guidelines to promote global consistency in regulatory processes, is also seeking to harmonize how AI is employed in drug development, ensuring safety, efficacy, and quality across international borders.[184]
Conclusion
Drug development has become increasingly complex, marked by longer timelines, higher risks, and rising costs. This complexity stems from several factors. First, scientists’ deepening understanding of biological processes related to health and disease requires more advanced research methods and technological tools. Second, stricter regulatory requirements demand extensive data collection and analysis to ensure safety and efficacy. Third, a growing emphasis on complex diseases, such as cancer and neurodegenerative disorders, requires innovative approaches, including new drug modalities. Fourth, increased efforts to diversify clinical trials have resulted in more comprehensive trial designs. Finally, the growing demand for precision medicine, which requires targeted approaches, further adds to this complexity.
Emerging technologies, particularly AI, hold the potential to transform biopharmaceutical innovation. AI can enhance various phases of drug development, from accelerating drug discovery and optimizing clinical trials to streamlining regulatory review and improving manufacturing and supply chains. These advances could significantly boost R&D productivity by enabling more efficient resource use, faster identification of viable drug candidates, and fewer costly clinical trial failures—ultimately fostering greater innovation and increasing the likelihood of bringing a higher number of effective, novel therapies to market sooner.
By expediting each stage of drug development, AI can accelerate access to breakthrough therapies, helping to meet urgent public health needs and reduce the disease burden on society. As illustrated in the case studies throughout this report, AI has a wide range of applications: It can improve inclusivity in clinical trials to ensure therapies are effective across diverse populations, thereby advancing the policy goal of health equity; when combined with EHRs, AI can help identify underdiagnosed individuals, particularly in underserved communities, reducing health disparities; in drug discovery, AI can accelerate therapeutic target identification and drug design; and in manufacturing, AI can enhance the design and production of gene therapies. Boosting research productivity, AI strengthens the biopharmaceutical sector’s contribution to innovation, promoting jobs, economic resilience, and global competitiveness. Moreover, AI can streamline regulatory processes by enhancing data quality and automating compliance, allowing for faster, safer access to essential therapies while maintaining rigorous safety standards.
A supportive public policy framework is key for the effective and responsible integration of AI in drug development. Key elements include public funding for basic research related to AI in drug development; educational initiatives to prepare the workforce for an AI-supported future in biopharmaceutical innovation; support for tools that enable privacy-enhancing sharing of and access to high-quality data; collaborations through efforts such as PPPs; and the development of a risk-based regulatory approach for evaluating AI tools, tailored to the level of risk each AI application may pose to patients. Such policies are essential to unlock AI’s full potential in drug development, accelerating the delivery of and broader access to potentially life-saving therapies.
Acknowledgments
The author would like to thank Robert Atkinson, Stephen Ezell, Daniel Castro, Hodan Omaar, Randolph Court, and Austin Slater for helpful feedback on this report. The author would also like to thank Asimov, Genentech, Gilead, and Johnson & Johnson for providing insights on the case studies. Any errors or omissions are the author’s sole responsibility.
About the Author
Sandra Barbosu, PhD, is associate director of ITIF’s Center for Life Sciences Innovation. Her research focuses on the economics of science and innovation, with a particular interest in emerging technologies in the health-care setting. Sandra is also adjunct professor in the Technology Management and Innovation Department at New York University's Tandon School of Engineering. She holds a PhD in Strategic Management from the Rotman School of Management at the University of Toronto, an MSc in Precision Cancer Medicine from the University of Oxford, and a BA in Economics and Mathematics from the University of Rochester.
About ITIF
The Information Technology and Innovation Foundation (ITIF) is an independent 501(c)(3) nonprofit, nonpartisan research and educational institute that has been recognized repeatedly as the world’s leading think tank for science and technology policy. Its mission is to formulate, evaluate, and promote policy solutions that accelerate innovation and boost productivity to spur growth, opportunity, and progress. For more information, visit itif.org/about.
Endnotes
[1]. Peter Saltonstall, Heidi Ross, and Paul T. Kim, “The Orphan Drug Act at 40: Legislative Triumph and the Challenges of Success,” The Milbank Quarterly, vol. 102, no. 1 (2024): 83–96, https://doi.org/10.1111/1468-0009.12680.
[2]. Elina Petrova, “Innovation in the pharmaceutical industry: The process of drug discovery and development,” Innovation and Marketing in the Pharmaceutical Industry: Emerging Practices, Research, and Policies (New York: Springer, 2013), 19–81.
[3]. David Proudman et al., “Public sector replacement of privately funded pharmaceutical R&D: cost and efficiency considerations,” Journal of Medical Economics, vol. 27, no. 1 (2024): 1253–1266, https://doi.org/10.1080/13696998.2024.2405407; Olivier J. Wouters, Martin McKee, Jeroen Luyten, “Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018,” JAMA, vol. 323, no. 9 (2020): 844–853, https://pmc.ncbi.nlm.nih.gov/articles/PMC7054832/.
[4]. “Clinical Development Success Rates and Contributing Factors 2011-2020,” Biotechnology Innovation Organization (February 2021), https://go.bio.org/rs/490-EHZ-999/images/ClinicalDevelopmentSuccessRates2011_2020.pdf.
[5]. “AI’s potential to accelerate drug discovery needs a reality check,” October 10, 2023, Editorial, Nature, 622: 217, https://doi.org/10.1038/d41586-023-03172-6.
[6]. Madura K.P. Jayatunga et al., “AI in small-molecule drug discovery: a coming wave?” Nature Reviews Drug Discovery, vol. 21, no. 3 (2022): 175–176, https://www.nature.com/articles/d41573-022-00025-1; Paul et al., “How to improve R&D productivity: the pharmaceutical industry's grand challenge.”
[7]. Dean G. Brown et al., “Clinical development times for innovative drugs,” Nature Reviews Drug Discovery, vol. 11 (2022): 793–794, https://www.nature.com/articles/d41573-021-00190-9.
[8]. “Drug development – The four phases,” Biostock Article Series, January 2, 2023, accessed July 11, 2024, https://www.biostock.se/en/2023/01/drug-development-the-four-phases/.
[9]. “Clinical Development Success Rates and Contributing Factors 2011-2020,” BIO.
[10]. Madura K.P. Jayatunga et al., “AI in small-molecule drug discovery: a coming wave?”; “Clinical Development Success Rates and Contributing Factors 2011-2020,” BIO.
[11]. Jessica Vamathevan et al., “Applications of machine learning in drug discovery and development,” Nature Reviews Drug Discovery, vol. 18 (2019): 463–477, https://www.nature.com/articles/s41573-019-0024-5.
[12]. Mingkun Lu et al., “Artificial Intelligence in Pharmaceutical Sciences,” Engineering, vol. 27 (2023): 37–69, https://www.sciencedirect.com/science/article/pii/S2095809923001649.
[13]. “2024 PhRMA Annual Membership Survey,” phrma.org, August 2024, https://phrma.org/-/media/Project/PhRMA/PhRMA-Org/PhRMA-Refresh/Report-PDFs/PhRMA_2024-Annual-Membership-Survey.pdf.
[14]. Ute Laermann-Nguyen and Martin Backfisch, “Innovation crisis in the pharmaceutical industry? A survey,” SN Business & Economics, vol. 1 (2021): 164, https://link.springer.com/article/10.1007/s43546-021-00163-5.
[15]. Laermann-Nguyen and Backfisch, “Innovation crisis in the pharmaceutical industry? A survey”; Compilation of CDER NME and New Biologic Approvals 1985-2022, U.S. Food and Drug Administration, March 13, 2023, https://www.fda.gov/drugs/drug-approvals-and-databases/compilation-cder-new-molecular-entity-nme-drug-and-new-biologic-approvals.
[16]. Deloitte Center for Health Solutions, “Unlocking R&D Productivity: Measuring the Return From Pharmaceutical Innovation 2018” (Deloitte Center for Health Solutions, 2018), https://www2.deloitte.com/global/en/pages/life-sciences-and-healthcare/articles/measuring-return-from-pharmaceutical-innovation.html.
[17]. Deloitte Center for Health Solutions, “Unlocking R&D Productivity: Measuring the Return From Pharmaceutical Innovation 2019” (Deloitte Center for Health Solutions, 2019), 2, https://www2.deloitte.com/content/dam/Deloitte/uk/Documents/life-sciences-health-care/deloitte-uk-ten-years-on-measuring-return-on-pharma-innovation-report-2019.pdf.
[18]. Deloitte Center for Health Solutions, “Ten years On: Measuring the return from pharmaceutical innovation 2019” (Deloitte Center for Health Solutions, 2019), 6, https://www2.deloitte.com/content/dam/Deloitte/uk/Documents/life-sciences-health-care/deloitte-uk-ten-years-on-measuring-return-on-pharma-innovation-report-2019.pdf.
[19]. Compilation of CDER NME and New Biologic Approvals 1985-2023, U.S. Food and Drug Administration; https://www.fda.gov/drugs/drug-approvals-and-databases/compilation-cder-new-molecular-entity-nme-drug-and-new-biologic-approvals.
[20]. Steven M. Paul et al., “How to improve R&D productivity: the pharmaceutical industry's grand challenge,” Nature Reviews Drug Discovery, vol. 9 (2010): 203–214, https://doi.org/10.1038/nrd3078.
[21]. Fabio Pammolli, Laura Magazzini, and Massimo Riccaboni, “The productivity crisis in pharmaceutical R&D,” Nature Reviews Drug Discovery, vol. 10 (2011): 428–438, https://www.nature.com/articles/nrd3405; Jack W. Scannell et al., “Diagnosing the decline in pharmaceutical R&D efficiency,” Nature Reviews Drug Discovery, vol. 11 (2012): 191200, https://www.nature.com/articles/nrd3681.
[22]. Bureau of Labor Statistics and Federal Reserve Economic Data, “Total Factor Productivity for Manufacturing: Pharmaceutical and Medicine Manufacturing (NAICS 3254) in the United States,” accessed December 14, 2023, https://fred.stlouisfed.org/series/IPUEN3254M001000000.
[23]. Bureau of Labor Statistics and Federal Reserve Economic Data, “Labor Productivity for Manufacturing: Pharmaceutical and Medicine Manufacturing (NAICS 3254) in the United States,” accessed December 14, 2023, https://fred.stlouisfed.org/series/IPUEN3254L001000000.
[24]. Joshua New, “The Promise of Data-Driven Drug Development” (ITIF, September 2019), https://www2.datainnovation.org/2019-data-driven-drug-development.pdf.
[25]. Mingkun Lu, et al., “Artificial Intelligence in Pharmaceutical Sciences.”
[26]. Ajay Agrawal, Joshua Gans, and Avi Goldfarb, Prediction Machines: The Simple Economics of Artificial Intelligence (Boston: Harvard Business Review Press), 2018.
[27]. Jayatunga et al., “AI in small-molecule drug discovery: a coming wave?”; Debleena Paul et al., “Artificial intelligence in drug discovery and development,” Drug Discovery Today, vol. 26, no. 1 (2021): 80–93, https://www.sciencedirect.com/science/article/pii/S1359644620304256?via%3Dihub.
[28]. Mingkun Lu, et al., “Artificial Intelligence in Pharmaceutical Sciences.”
[29]. Jessica Vamathevan et al., “Applications of machine learning in drug discovery and development.”
[30]. Kit-Kay Mak, Yi-Hang Wong, and Mallikarjuna Rao Pichika, “Artificial Intelligence in Drug Discovery and Development,” In: F. J. Hock and M. K. Pugsley (eds) Drug Discovery and Evaluation: Safety and Pharmacokinetic Assays, Springer, Cham (2023): 1–38, https://doi.org/10.1007/978-3-030-73317-9_92-1.
[31]. “AlphaFold Protein Structure Database,” EMBL-EBI, accessed July 12, 2024, https://alphafold.ebi.ac.uk.
[32]. “Method of the year 2021: Protein structure prediction,” Nature Methods, vol. 19, no. 1 (2022), https://doi.org/10.1038/s41592-021-01380-4.
[33]. “The Nobel Prize in Chemistry 2024: They cracked the code for proteins’ amazing structures,” The Royal Swedish Academy of Sciences, October 9, 2024, https://www.kva.se/en/news/the-nobel-prize-in-chemistry-2024/.
[34]. Bin Feng et al, “A bioactivity foundation model using pairwise meta-learning,” Nature Machine Intelligence, vol. 6 (2024): 962–974, https://www.nature.com/articles/s42256-024-00876-w.
[35]. “Genentech, NVIDIA partner on ‘lab-in-a-loop’ AI platform,” PharmaPhorum, November 21, 2023, https://pharmaphorum.com/news/genentech-nvidia-partner-lab-loop-ai-platform; “Redefining Drug Discovery with AI,” Genentech, March 18, 2024, https://www.gene.com/stories/redefining-drug-discovery-with-ai.
[36]. “Genentech and NVIDIA Revolutionize Drug Discovery with Generative AI and Lab in the Loop,” NVIDIA YouTube Channel, November 21, 2023, https://www.youtube.com/watch?v=-Ijg2g8AsjE.
[37]. Madura KP Jayatunga, et al., “AI in small-molecule drug discovery: a coming wave?”; Paul et al., “How to improve R&D productivity: the pharmaceutical industry’s grand challenge.”
[38]. “Unlocking the Potential of AI in Drug Discovery: Current status, barriers and future opportunities,” Wellcome–BCG, June 2023, https://cms.wellcome.org/sites/default/files/2023-06/unlocking-the-potential-of-AI-in-drug-discovery_report.pdf.
[39]. “Artificial Intelligence in Health Care,” U.S. Government Accountability Office and National Academy of Medicine Report, December 2019, https://www.gao.gov/assets/gao-20-215sp.pdf.
[40]. Alex Keown “An AI First: Deep Genomics Platform Reveals Wilson’s Disease Drug Candidate,” September 26, 2019, https://www.biospace.com/deep-genomics-ai-program-reveals-first-drug-candidate-for-wilson-s-disease.
[41]. Ibid.
[42]. Talha Burki, “A new paradigm for drug development,” Lancet Digital Health, vol. 2, no. 5 (2020): E226-E227, https://doi.org/10.1016%2FS2589-7500(20)30088-1.
[43]. “First drug discovered and designed with generative AI enters Phase II trials, with first patients dosed,” EurekAlert! AAAS Business Announcement, Insilico Medicine, June 27, 2023, https://www.eurekalert.org/news-releases/993844; “New Milestone in AI Drug Discovery,” Insilico Medicine, July 1 2023, https://insilico.com/blog/first_phase2.
[44]. Andrew Dunn, “After years of hype, the first AI-designed drugs fall short in the clinic,” Endpoints in Focus, Endpoints News, October 19, 2023, https://endpts.com/first-ai-designed-drugs-fall-short-in-the-clinic-following-years-of-hype/.
[45]. Madura K. P. Jayatunga et al., “AI in small-molecule drug discovery: a coming wave?”
[46]. Ibid.
[47]. “Artificial Intelligence for Drug Discovery 2023 Teaser,” Deep Pharma Intelligence (2023) https://analytics.dkv.global/deep-pharma/Teaser-AI-DD.pdf.
[48]. Ibid.
[49]. Ibid.
[50]. “AI in Pharmaceuticals Promises Innovation, Speed, and Savings,” S&P Global Ratings, October 1, 2024, https://www.spglobal.com/ratings/en/research/articles/241001-ai-in-pharmaceuticals-promises-innovation-speed-and-savings-13254002.
[51]. “Artificial Intelligence for Drug Discovery 2023 Teaser.”
[52]. Junyu Luo et al., “ClinicalRisk: A New therapy-related Clinical Trial Dataset for Predicting Trial Status and Failure Reasons,” Proceedings of the 32nd ACM International Conference on Information and Knowledge Management (October 2023): 5356–5360, 10.1145/3583780.3615113.
[53]. Yingzhou Lu et al., “Uncertainty quantification and interpretability for clinical trial approval prediction,” Health Data Science, vol. 4 (2024): 0126, https://arxiv.org/pdf/2401.03482; “Clinical trials market size, share & trends analysis report by phase (phase I, phase ii, phase iii, phase iv), by study design (interventional, observational, expanded access), by indication, by region, and segment forecasts 2021-2028,” Grand View Research (2021), https://www.grandviewresearch.com/industry-analysis/clinical-trial-kits-market-report.
[54]. Dean G. Brown et al., “Clinical development times for innovative drugs,” Nature Reviews Drug Discovery, vol. 21 (2022): 793–794, https://www.nature.com/articles/d41573-021-00190-9.
[55]. “Clinical Development Success Rates and Contributing Factors, 2011-2020,” Biotechnology Innovation Organization, Informa Pharma Intelligence, and QLS Advisors (February 2021) https://www.bio.org/clinical-development-success-rates-and-contributing-factors-2011-2020.
[56]. Ibid; Paul et al., “How to improve R&D productivity: the pharmaceutical industry’s grand challenge.”
[57]. Matthew Hutson, “How AI is being used to accelerate clinical trials,” Nature Index, March 13, 2024, https://doi.org/10.1038/d41586-024-00753-x.
[58]. Ibid.
[59]. Ruishan Liu et al., “Evaluating eligibility criteria of oncology trials using real-world data and AI,” Nature, vol. 592 (2021): 629-633, https://doi.org/10.1038/s41586-021-03430-5.
[60]. Ibid.
[61]. Ryan D. Nipp, Kessely Hong, and Electra D. Paskett, “Overcoming Barriers to Clinical Trial Enrollment,” American Society of Clinical Oncology Educational Book, vol. 39 (2019), https://ascopubs.org/doi/full/10.1200/EDBK_243729.
[62]. Yilu Fang et al., “Combining human and machine intelligence for clinical trial eligibility querying,” Journal of the American Medical Informatics Association, vol. 29, no. 7 (2022): 1161–1171, https://academic.oup.com/jamia/article/29/7/1161/6569054.
[63]. Jimyung Park et al., “Criteria2Query 3.0: Leveraging generative large language models for clinical trial eligibility query generation,” Journal of Biomedical Informatics, vol. 154, no. C (2024), https://dl.acm.org/doi/10.1016/j.jbi.2024.104649.
[64]. Qiao Jin et al., “Matching patients to clinical trials with large language models,” ArXiv [Preprint], April 2024, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10418514/.
[65]. “How Our Llama Grant Recipients Are Tackling Global Issues,” Meta, September 24, 2024, https://about.fb.com/news/2024/09/llama-grant-recipients-are-tackling-global-issues/.
[66]. Ibid.
[67]. Arsen Osipov et al., “The Molecular Twin artificial-intelligence platform integrates multi-omic data to predict outcomes for pancreatic adenocarcinoma patients,” Nature Cancer, vol. 5 (2024): 299-314, https://www.nature.com/articles/s43018-023-00697-7.
[68]. “Using AI to Create a Better Test to Predict Pancreatic Cancer Progression,” Let’s Win Pancreatic Cancer, May 10, 2024, https://letswinpc.org/research/ai-find-progression-biomarkers/.
[69]. Tianfan Fu et al., “HINT: Hierarchical interaction network for clinical-trial-outcome predictions,” Patterns, vol. 3 no. 4 (2022), https://doi.org/10.1016%2Fj.patter.2022.100445.
[70]. Zifeng Wang, Cao Xiao, and Jimeng Sun, “SPOT: sequential predictive modeling of clinical trial outcome with meta-learning,” Proceedings of the 14th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, 53: 1–11, https://doi.org/10.1145/3584371.3613001.
[71]. Matthew Hutson, “How AI is being used to accelerate clinical trials”; Hunki Paek et al., “Granularly, Precisely, and Timely: Leveraging Large Language Models for Safety and Efficacy Extraction in Oncology Clinical Trial Abstracts (SEETrials),” medRxi [Preprint], May 2024, https://www.medrxiv.org/content/10.1101/2024.01.18.24301502v1.
[72]. Jasmine Pennis, “QuantHealth Launches Katina: AI-Powered Platform for Clinical Trial Optimization,” HIT Consultant, January 12, 2023, https://hitconsultant.net/2023/12/01/quanthealth-launches-katina-ai-powered-platform-for-clinical-trial-optimization/.
[73]. Brian Buntz, “QuantHealth’s AI simulates 100+ clinical trials with 85% accuracy,” Drug Discovery & Development, August 23, 2024, https://www.drugdiscoverytrends.com/quanthealths-ai-simulates-100-clinical-trials-with-85-accuracy/.
[74]. “Protocol Registration Quality Control Review Criteria,” ClinicalTrials.gov, June 7, 2024, accessed July 31, 2024, https://clinicaltrials.gov/submit-studies/prs-help/protocol-registration-quality-control-review-criteria.
[75]. Team Snorkel, “Augmenting the clinical trial design process with information extraction,” Snorkel.ai, February 22, 2022, https://snorkel.ai/augmenting-the-clinical-trial-design-information-extraction/; https://www.snorkel.org/get-started/.
[76]. “2020 Drug Trials Snapshots Summary Report,” U.S. Food and Drug Administration (2020), https://www.fda.gov/media/145718/download.
[77]. Carol Cruickshank, Cian Wade, and Junaid Bajwa, “How AI Could Help Reduce Inequities in Health Care,” Harvard Business Review, August 29, 2024, https://hbr.org/2024/08/how-ai-could-help-reduce-inequities-in-health-care.
[78]. Angel CY Mak et al., “Whole-Genome Sequencing of Pharmacogenetic Drug Response in Racially Diverse Children with Asthma,” American Journal of Respiratory and Critical Care Medicine, vol. 197, no. 12 (2018): 1552–1564, https://doi.org/10.1164/rccm.201712-2529OC.
[79]. “The Power of Diversity in Clinical Trials,” Novavax Contributor, Forbes, May 6, 2023, https://www.forbes.com/sites/novavax/2023/03/06/the-power-of-diversity-in-clinical-trials/.
[81]. Bindu Kanapuru et al., “Analysis of racial and ethnic disparities in multiple myeloma US FDA drug approval trials,” Blood Advances, vol. 6, no. 6 (2022): 1684–1691, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8941450/.
[82]. Ruchika S. Patil, Samruddhi B. Kulkarni, and Vinod L. Gaikwad, “Artificial intelligence in pharmaceutical regulatory affairs,” Drug Discovery Today (2023): 103700, https://doi.org/10.1016/j.drudis.2023.103700.
[83]. “Unlock the true power of clinical trial data,” Medidata, accessed July 10, 2024, https://www.medidata.com/wp-content/uploads/2024/08/Clinical-Data-Studio-Infographic-Jul-24-2.pdf.
[84]. “Regulatory Transformation with Unified RIM,” Veeva, 2024, https://www.veeva.com/products/vault-rim/.
[85]. “Veeva Vault Submissions,” Veeva, 2024, https://www.veeva.com/products/vault-submissions/.
[86]. Sudeep Srivastasa, “How AI in Demand Forecasting is Enhancing Supply Chain Efficiency,” August 28, 2024, AppInventiv, https://appinventiv.com/blog/ai-for-demand-forecasting/; “Generative AI in the pharmaceutical industry: Moving from hype to reality,” McKinsey & Company Report, January 9, 2024, https://www.mckinsey.com/industries/life-sciences/our-insights/generative-ai-in-the-pharmaceutical-industry-moving-from-hype-to-reality.
[87]. “AI-Designed Promoters,” Asimov, June 20, 2023, https://blog.asimov.com/blog-post/ai-promoters.
[88]. “Drug Shortages,” Pew Trusts, January 10, 2017, https://www.pewtrusts.org/en/research-and-analysis/reports/2017/01/drug-shortages.
[89]. “AI could strengthen pharmaceutical supply chains to save lives,” IT-Online, August 19, 2024, https://it-online.co.za/2024/08/19/ai-could-strengthen-pharmaceutical-supply-chains-to-save-lives/
[90]. Ibid.
[91]. Clint Boulton, “Merck prescribes AI for supply chain complexity,” CIO, December 17, 2019, https://www.cio.com/article/215839/merck-prescribes-ai-for-supply-chain-complexity.html.
[92]. Avi Goldfarb and Florenta Teodoridis, “Why is AI adoption in health care lagging?” Brookings Institution (March 2022), https://www.brookings.edu/articles/why-is-ai-adoption-in-health-care-lagging/.
[93]. Alexandre Blanco-Gonzalez et al., “The Role of AI in Drug Discovery: Challenges, Opportunities, and Strategies,” Pharmaceuticals, vol. 16, no. 6 (2023): 891, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10302890/.
[94]. Kendall Powell, “The broken promise that undermines human genome research,” Nature News Feature, February 10, 2021, https://www.nature.com/articles/d41586-021-00331-5; International Human Genome Sequencing Consortium, “Initial sequencing and analysis of the human genome,” Nature, vol. 409 (2001): 8600921, https://www.nature.com/articles/35057062; Francis S. Collins and Leslie Fink, “The Human Genome Project,” Alcohol Health Research World, vol. 19, no.3 (1995): 190195, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6875757/.
[95]. “The Bermuda Principles,” Duke University Libraries, accessed September 22, 2024, https://dukespace.lib.duke.edu/communities/9bf2e54b-6912-407e-90a7-22213ff0c90a.
[96]. Powell, “The broken promise that undermines human genome research.”
[97]. Ibid.
[98]. “Genome-Wide Association Studies (GWAS),” National Human Genome Research Institute, September 22, 2024, https://www.genome.gov/genetics-glossary/Genome-Wide-Association-Studies.
[99]. Powell, “The broken promise that undermines human genome research.”
[100]. “Unlocking the power of genomic data to benefit human health,” Global Alliance for Genomics & Health, accessed July 20, 2024, https://www.ga4gh.org.
[101]. Annalisa Buniello et al., “The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019,” Nucleic Acids Research, vol. 47, no. D1 (2019): D1005-D1012, https://academic.oup.com/nar/article/47/D1/D1005/5184712?login=false; “GWAS Catalog,” European Bioinformatics Institute (EMBL-EBI) and National Human Genome Research Institute (NHGRI), accessed July 20, 2024, https://www.ebi.ac.uk/gwas/home; Vamathevan et al., “Applications of machine learning in drug discovery and development.”
[102]. “Human Cell Atlas,” accessed July 24, 2024, https://www.humancellatlas.org.
[103]. “H.R.1 – American Recovery and Reinvestment Act of 2009,” 111th Congress 2009–2010, accessed August 1, 2024, http://www.congress.gov/bill/111th-congress/house-bill/1.
[104]. “National Trends in Hospital and Physician Adoption of Electronic Health Records,” HealthIT.gov, accessed August 20, 2024, https://www.healthit.gov/data/quickstats/national-trends-hospital-and-physician-adoption-electronic-health-records.
[105]. Maggy Bobek Tieche, “Most common hospital HER systems by market share,” Definitive Healthcare, January 10, 2024, https://www.definitivehc.com/blog/most-common-inpatient-ehr-systems.
[106]. Miriam Reisman, “The Challenge of Making Electronic Data Usable and Interoperable,” Pharmacy & Therapeutics, vol. 42, no. 9 (2017): 572-575, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5565131/.
[107]. Zhaoyi Chen et al., “Applications of artificial intelligence in drug development using real-world data,” Drug Discovery Today, vol. 26. no. 5 (2021): 1256–1264, https://doi.org/10.1016%2Fj.drudis.2020.12.013.
[108]. Ibid.
[110]. Charles Friedman et al., “Toward a science of learning systems: a research agenda for the high-functioning Learning Health System,” Journal of the American Medical Informatics Association, vol. 22, no. 1 (2015): 43–50, https://academic.oup.com/jamia/article/22/1/43/834511.
[111]. Zhaoyi Chen et al., “Applications of artificial intelligence in drug development using real-world data.”
[112]. Isaac S. Kohane, “Using electronic health records to drive discovery in disease genomics,” Nature Reviews Genetics, vol. 12 (2011): 417-428, https://www.nature.com/articles/nrg2999.
[113]. Ibid.
[114]. Zoom call with Lucia Savage, chief privacy and regulatory officer at Omada Health and former chief privacy officer at U.S. Department of Health and Human Services Office of the National Coordinator for Health IT, September 9, 2024; Laura Bradford, Mateo Aboy, and Kathleen Liddell, “International transfers of health data between the EU and USA: a sector-specific approach for the USA to ensure an ‘adequate’ level of protection,” Journal of Law and the Biosciences, vol. 7, no. 1 (2020), https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8249089/.
[115]. John Rigg et al., “Finding undiagnosed patients with hepatitis C virus: an application of machine learning to US ambulatory electronic medical records,” BMJ Health & Care Informatics, vol. 20, no. 1 (2023): e100651, https://pubmed.ncbi.nlm.nih.gov/36639190/.
[116]. “Synthetic Data in Drug Development – What it is and How it Relates to AI-informed Approaches,” VERISIMLife, May 19, 2023, https://www.verisimlife.com/publications-blog/synthetic-data-in-drug-development-what-it-is-and-how-it-relates-to-ai-informed-approaches.
[117]. “Innovative Health Initiative,” https://www.ihi.europa.eu; “Alzheimer’s Disease Neuroimaging Initiative,” https://adni.loni.usc.edu; “Project Data Sphere,” https://data.projectdatasphere.org/projectdatasphere/html/home; “Open Targets,” https://www.opentargets.org; “Structural Genomics Consortium,” https://www.thesgc.org.
[118]. “Flatiron Health,” https://flatiron.com.
[119]. Aljosa Smajic, Melanie Grandits, Gerhard F. Ecker, ”Privacy-preserving techniques for decentralized and secure machine learning in drug discovery,” Drug Discovery Today, vol. 28, no. 12 (2023): 103820, https://www.sciencedirect.com/science/article/pii/S1359644623003367#b0040.
[120]. Sandra Barbosu, “Advancing Biomedical Innovation with Policies Supporting Privacy-Enhancing Technologies” (ITIF, May 2024), https://itif.org/publications/2024/05/13/advancing-biomedical-innovation-with-privacy-enhancing-technologies/.
[121]. Smajic et al., “Privacy-preserving techniques for decentralized and secure machine learning in drug discovery.”
[122]. Ibid.
[123]. Rong Ma et al., “Secure multiparty computation for privacy-preserving drug discovery,” Bioinformatics, vol. 36, no. 9 (2020): 2872-2880, https://academic.oup.com/bioinformatics/article/36/9/2872/5709032; Hyunghoon Cho, David J. Wu and Bonnier Berger, “Secure genome-wide association analysis using multiparty computation,” Nature Biotechnology, vol. 36, no. 5 (2018): 547-551, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5990440/; Brian Hie, Hyunghoon Cho, and Bonnier Berger, “Realizing private and practical pharmacological collaboration,” Science, vol. 362, no. 6412 (2018): 347–350, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6519716/.
[124]. Smajic et al., “Privacy-preserving techniques for decentralized and secure machine learning in drug discovery.”
[125]. Cloud Hacks, “Federated Learning: A Paradigm Shift in Data Privacy and Model Training,” March 1, 2024, https://medium.com/@cloudhacks_/federated-learning-a-paradigm-shift-in-data-privacy-and-model-training-a41519c5fd7e.
[126]. “MELLODDY: A ‘co-opetitive’ machine learning platform powered by Owkin,” February 17, 2020, https://www.owkin.com/blogs-case-studies/melloddy-a-co-opetitive-platform-for-machine-learning-across-companies-powered-by-owkin-technology.
[127]. Craig Gentry, “Computing arbitrary functions of encrypted data,” Communications of the ACM, vol. 53, no. 3 (2010): 97–105, https://dl.acm.org/doi/abs/10.1145/1666420.1666444.
[128]. “Homomorphic Encryption Standardization,” https://homomorphicencryption.org/standard/.
[129]. “Encrypted Molecular Discovery using FHE,” May 2, 2024, https://medium.com/@janweinreich286/encrypted-molecular-discovery-using-fully-homomorphic-encryption-069c45ee152e.
[130]. Smajic et al., “Privacy-preserving techniques for decentralized and secure machine learning in drug discovery.”
[131]. Cynthia Dwork, “Differential Privacy,” In: Michele Bugliesi et al. (Eds.), Automata, Languages and Programming, ICALP 2006, Lecture Notes in Computer Science, vol. 4052, Springer, Berlin, Heidelberg, https://link.springer.com/chapter/10.1007/11787006_1.
[132]. Md. Mohaiminul Islam et al., “Differential private deep learning models for analyzing breast cancer omics data,” Frontiers in Oncology, vol. 12 (2022): 879607, https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2022.879607/full; Smajic et al., “Privacy-preserving techniques for decentralized and secure machine learning in drug discovery.”
[133]. “What is differential privacy,” OpenDP, https://docs.opendp.org/en/stable/index.html; Duality Technologies, https://dualitytech.com; “DataSafes,” Actuate, https://actuateinnovation.org/programs/datasafes/; OpenMined, https://openmined.org.
[134]. “Duality Technologies joins the fight against cancer with a new secure data collaboration toolkit enabling analytics and AI on Oncological Real-World Data,” PR NewsWire, August 15, 2023, https://www.prnewswire.com/news-releases/duality-technologies-joins-the-fight-against-cancer-with-a-new-secure-data-collaboration-toolkit-enabling-analytics-and-ai-on-oncological-real-world-data-301901187.html.
[135]. Sajid Ali et al., “Explainable Artificial Intelligence (XAI): What we know and what is left to attain Trustworthy Artificial Intelligence,” Information Fusion, vol. 99 (2023), 101805, https://doi.org/10.1016/j.inffus.2023.101805.
[136]. Roohallah Alizadehsani et al., “Explainable Artificial Intelligence for Drug Discovery and Development – A Comprehensive Survey,” IEEE Access (2024), https://arxiv.org/pdf/2309.12177.
[137]. Jose Jimenez-Luna, Francesca Grisoni, and Gisbert Schneider, “Drug discovery with explainable artificial intelligence,” Nature Machine Intelligence, vol. 2 (2020): 573–584, https://www.nature.com/articles/s42256-020-00236-4.
[138]. “3: AI Risks and Trustworthiness,” NIST, accessed August 20, 2024, https://airc.nist.gov/AI_RMF_Knowledge_Base/AI_RMF/Foundational_Information/3-sec-characteristics.
[139]. Avi Goldfarb and Florenta Teodoridis, “Why is AI adoption in health care lagging?”; AI Now Institute, “AI Now 2019 Report,” (December 2019), https://ainowinstitute.org/AI_Now_2019_Report.pdf; National Science Foundation, Award Abstract #2229885, Institute for Trustworthy AI in Law and Society (TRAILS), https://www.nsf.gov/awardsearch/showAward?AWD_ID=2229885&HistoricalAwards=false.
[140]. “Machine Learning Algorithmic Bias,” FDA Digital Health and Artificial Intelligence Glossary – Educational Resource, https://www.fda.gov/science-research/artificial-intelligence-and-medical-products/fda-digital-health-and-artificial-intelligence-glossary-educational-resource.
[141]. Anna C. Need and David B. Goldstein, “Next generation disparities in human genomics: concerns and remedies,” Trends in Genetics, vol. 25, no.11 (2009): 489–494, https://www.cell.com/trends/genetics/abstract/S0168-9525(09)00185-1; Giorgio Sirugo, Scott M. Williams, and Sarah A. Tishkoff, “The Missing Diversity in Human Genetic Studies,” Cell, vol. 177, no. 1 (2019): 26–31, https://www.cell.com/cell/fulltext/S0092-8674(19)30231-4?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0092867419302314%3Fshowall%3Dtrue&.
[142]. “Why Diverse Representation in Clinical Research Matters and the Current State of Representation within the Clinical Research Ecosystem” in Improving Representation in Clinical Trials and Research: Building Research Equity for Women and Underrepresented Groups (2022): https://www.ncbi.nlm.nih.gov/books/NBK584396/.
[143]. Alice B. Popejoy and Stephanie M. Fullerton, “Genomics is failing on diversity,” Nature, vol. 538 (2016): 161–164, https://www.nature.com/articles/538161a.
[144]. Isaac S. Kohane, “Using electronic health records to drive discovery in disease genomics.”
[145]. Diana W. Bianchi et al., “The All of Us Research Program is an opportunity to enhance the diversity of US biomedical research,” Nature Medicine, vol. 30 (2024): 330–333, https://www.nature.com/articles/s41591-023-02744-3.
[146]. Ewen Callaway, “World’s biggest set of human genome sequences opens to scientists,” Nature News, November 30, 2023, https://www.nature.com/articles/d41586-023-03763-3.
[147]. Ken Suzuki et al., “Genetic drivers of heterogeneity in type 2 diabetes pathophysiology,” Nature, vol. 627, no. 8003 (2024): 347–357, https://www.nature.com/articles/s41586-024-07019-6.pdf; Popejoy and Fullerton, “Genomics is failing on diversity.”
[148]. Neil Savage, “Why artificial intelligence needs to understand consequences,” Nature Outlook, February 24, 2023, https://www.nature.com/articles/d41586-023-00577-1; “The AI Index Report 2024,” Stanford University Institute for Human-Centered Artificial Intelligence (2024), https://aiindex.stanford.edu/report/.
[149]. Avi Goldfarb and Florenta Teodoridis, “Why is AI adoption in health care lagging?”
[150]. Ajay Agrawal et al., Prediction Machines: The Simple Economics of Artificial Intelligence; Erik Brynjolfsson, Tom Mitchell, and Daniel Rock, “What Can Machines Learn, and What Does It Mean for Occupations and the Economy?” AEA Papers and Proceedings, vol. 108 (2018), https://www.aeaweb.org/articles?id=10.1257/pandp.20181019.
[151]. “Scientists ‘cautiously optimistic’ about AI’s role in drug discovery,” Phys.org, August 2, 2024, https://phys.org/news/2024-08-scientists-cautiously-optimistic-ai-role.html; Xinyu Gu, Akashnathan Aranganathan, and Pratyush Tiwary, “Empowering AlphaFold2 for protein conformation selective drug discovery with AlphaFold2-RAVE,” [Preprint] (2024), https://elifesciences.org/reviewed-preprints/99702v1.
[152]. Avi Goldfarb and Florenta Teodoridis, “Why is AI adoption in health care lagging?”
[153]. New, “The Promise of Data-Driven Drug Development.”
[154]. Ron Southwick, “TEFCA is up and running. Here’s why it’s a big deal for the exchange of health data,” Chief Healthcare Executive, December 14, 2023, https://www.chiefhealthcareexecutive.com/view/tefca-is-up-and-running-here-s-why-it-s-a-big-deal-for-the-exchange-of-health-data-; “TEFCA,” Sequoia Project, https://rce.sequoiaproject.org/tefca/.
[155]. “New data exchange network shows potential to strengthen public health,” Pew Trust, July 12, 2024, https://www.pewtrusts.org/en/research-and-analysis/articles/2024/07/12/new-data-exchange-network-shows-potential-to-strengthen-public-health.
[156]. “HHS Expands TEFCA by adding two additional QHINs,” U.S. Department of Health and Human Services, February 12, 2024, https://www.hhs.gov/about/news/2024/02/12/hhs-expands-tefca-by-adding-two-additional-qhins.html.
[157]. “H.R.5116 – Behavioral Health Information Technology Coordination Act,” 118th Congress 2023–2024, https://www.congress.gov/bill/118th-congress/house-bill/5116/text.
[158]. New, “The Promise of Data-Driven Drug Development.”
[159]. “Executive Order on the safe, secure, and trustworthy development and use of artificial intelligence,” The White House, October 30, 2023, https://www.whitehouse.gov/briefing-room/presidential-actions/2023/10/30/executive-order-on-the-safe-secure-and-trustworthy-development-and-use-of-artificial-intelligence/.
[160]. “H.R.4755 – Privacy Enhancing Technology Research Act,” 118th Congress 2023–2024, https://www.congress.gov/bill/118th-congress/house-bill/4755.
[161]. “Duality Technologies joins the fight against cancer with a new secure data collaboration toolkit enabling analytics and AI on Oncological Real-World Data.”
[162]. New, “The Promise of Data-Driven Drug Development”; Barbosu, “Advancing Biomedical Innovation with Policies Supporting Privacy-Enhancing Technologies.”
[163]. “Cancer Moonshot,” NIH National Cancer Institute, https://www.cancer.gov/research/key-initiatives/moonshot-cancer-initiative.
[164]. Remco L. A. de Vrueh and Daan J. A. Crommelin, “Reflections on the Future of Pharmaceutical Public-Private Partnerships: From Input to Impact,” Pharmaceutical Research, vol. 34 (2017): 1985–1999, https://link.springer.com/article/10.1007/s11095-017-2192-5#Sec12; Smajic et al., “Privacy-preserving techniques for decentralized and secure machine learning in drug discovery.”
[165]. Open Targets, https://www.opentargets.org.
[166]. Structural Genomics Consortium, https://www.thesgc.org.
[167]. Conscience, https://conscience.ca/about-us/.
[168]. de Vrueh and Crommelin, “Reflections on the Future of Pharmaceutical Public-Private Partnerships: From Input to Impact.”
[169]. “Bridge2AI,” NIH, https://commonfund.nih.gov/bridge2ai.
[170]. National Science Foundation Award Abstract #2409105, NSF SBIR Phase I: Development of an AI-driven humanized and developable single-domain library design platform for accelerated drug discovery,” July 1, 2024, https://www.nsf.gov/awardsearch/showAward?AWD_ID=2409105.
[171]. “ARPA-H leverages AI to transform healthcare technology and outcomes,” Health HQ, June 2024, https://healthhq.world/issue-sections/healthcare-technology/healthcare-and-cybersecurity/arpa-h-leverages-ai-to-transform-healthcare-technology-and-outcomes/.
[172]. “Accelerating Medicines Partnership,” NIH, https://www.nih.gov/research-training/accelerating-medicines-partnership-amp.
[173]. “Bridge2AI,” NIH, https://commonfund.nih.gov/bridge2ai.
[174]. “National Artificial Intelligence Research Institutes,” National Science Foundation, https://new.nsf.gov/funding/opportunities/national-artificial-intelligence-research-institutes.
[175]. “Big Data to Knowledge,” NIH, https://commonfund.nih.gov/bd2k.
[176]. New, “The Promise of Data-Driven Drug Development.”
[177]. Zachary Arnold et al., “Immigration Policy and the U.S. AI Sector,” Center for Security and Emerging Technology, September 2019, https://cset.georgetown.edu/publication/immigration-policy-and-the-u-s-ai-sector/.
[178]. “Executive Order on the safe, secure, and trustworthy development and use of artificial intelligence” The White House, October 30, 2023.
[179]. “FDA releases two discussion papers to spur conversation about AI and ML in drug development and manufacturing,” US FDA, https://www.fda.gov/news-events/fda-voices/fda-releases-two-discussion-papers-spur-conversation-about-artificial-intelligence-and-machine.
[180]. “The use of AI in the medicinal product lifecycle,” EU EMA (2023), https://www.ema.europa.eu/en/use-artificial-intelligence-ai-medicinal-product-lifecycle.
[181]. Ibid; Sarah Cowlishaw and Ellie Handy, “EMA releases reflection paper on AI/ML in the medicinal product lifecycle, Covington, Jule 21, 2023, https://www.insideeulifesciences.com/2023/07/21/ema-releases-reflection-paper-on-ai-ml-in-the-medicinal-product-lifecycle/.
[182]. “Using Artificial Intelligence & Machine Learning in the Development of Drug & Biological Products: Discussion Paper and Request for Feedback,” U.S. FDA, May 2023, https://www.fda.gov/media/167973/download.
[183]. Joanne S. Eglovitch, “FDA plans to release AI drug development guidance this year,” May 30, 2024, Regulatory Focus, https://www.raps.org/news-and-articles/news-articles/2024/5/fda-plans-to-release-ai-drug-development-guidance.
[184]. International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use, https://www.ich.org/.
Editors’ Recommendations
September 18, 2019


